| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,340,933 – 13,341,047 |
| Length | 114 |
| Max. P | 0.576794 |

| Location | 13,340,933 – 13,341,047 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
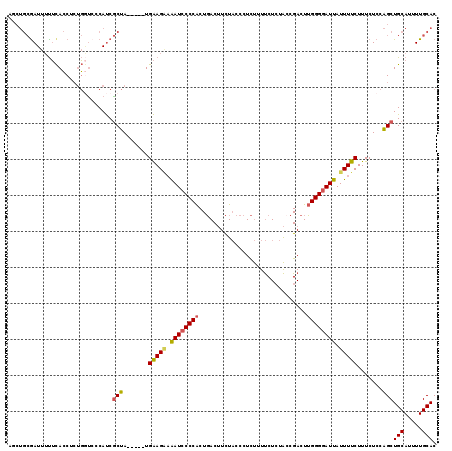
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.40 |
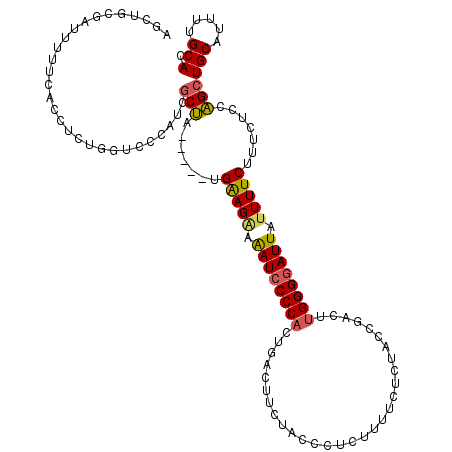
| Structure conservation index | 0.66 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13340933 114 + 23771897 AGCUGCGAUUUUUCACCUCUGGUCCCAUCGCUA-----UGAAGGAAAUCCCCACUGACUUCUACCCUCUUUUCUCUACCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC (((((((((.....(((...)))...)))))..-----.((((((((((((((..............................)))))))....)))))))...))))((.....)).. ( -24.51) >DroSec_CAF1 12777 114 + 1 AGCUGCGAUUUUUCACCCCUGGUCCCAUCGCUA-----UGAAGAAAAUCCCCACUGACUUCUACCCUCUUUUCUCUACCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC (((((((((.....(((...)))...)))))..-----.(((((.((((((((..............................)))))))).))))).......))))((.....)).. ( -24.21) >DroSim_CAF1 12344 114 + 1 AGCUGCAAUUUUUCACCCCUGGUCCCAUCGCUA-----UGAAGAAAAUCCCCACUGACUUCUACCCUCUUUUCUCUACCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC (((((..............((((......))))-----.(((((.((((((((..............................)))))))).)))))......)))))((.....)).. ( -21.41) >DroEre_CAF1 12942 113 + 1 UCCUGCGAUU-UUCACCUCCGCUCCCAUCGCUA-----UGAAGAAAAUCCCCACUGACUUCUACCCUCUUUUCUCUACCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC ...(((((..-(.((.((..((.......))..-----.(((((.((((((((..............................)))))))).))))).......)).)).)..))))). ( -19.01) >DroYak_CAF1 13405 114 + 1 UUCUGCGAUUUUUCACCUCUGGCGGCAUCGCUA-----UGAAGGAAAUCCCCACUGACUUCUACCCUCUUUUCUCUAGCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC ...(((((....)).......(((((.((((((-----.((.(((((........(....).......)))))))))))))..((((((..........))))))))))).....))). ( -26.96) >DroAna_CAF1 11860 108 + 1 ------GAUUUCAUAGCUAUUAUCUCACCACCAGCAGCUGGAG--GAUACCCGCUCACAUUU--CCUUCGUCCU-CGUCGACUGGGGGAUUAUUUUCUUUCUCCGGCUGCAUUUUGCAC ------...........................((((((((((--((.............((--(((..(((..-....)))..))))).........))))))))))))......... ( -29.65) >consensus AGCUGCGAUUUUUCACCUCUGGUCCCAUCGCUA_____UGAAGAAAAUCCCCACUGACUUCUACCCUCUUUUCUCUACCGACUUGGGGAUUAUUUUCUUUCUCCAGCUGCAUUUUGCAC .............................(((.......(((((.((((((((..............................)))))))).))))).......)))(((.....))). (-15.99 = -15.93 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:03 2006