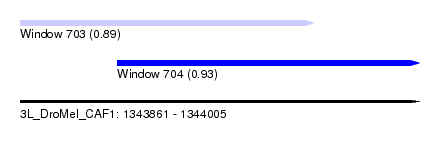
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,343,861 – 1,344,005 |
| Length | 144 |
| Max. P | 0.930962 |

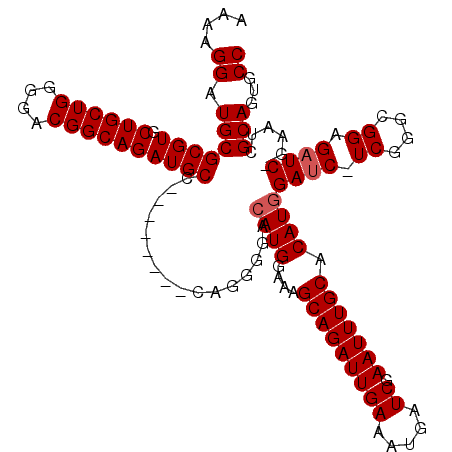
| Location | 1,343,861 – 1,343,967 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -27.56 |
| Energy contribution | -28.56 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1343861 106 + 23771897 C-----GAGAGUCCUGAGUCCUUUUGUCCUGGUGUUUACAAAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGC---------CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUU .-----..(..(((...(((((((.(((((..((....))...)))))(.((((.((((((....))))))))))---------))))))))..)))...).((((.....))))..... ( -36.80) >DroSec_CAF1 16114 105 + 1 U-----GAGAAUCCCGAGUCCU-UUGUCCUGGUGUUUACAAAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGC---------CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUU .-----.......(((.(((((-(((((((..((....))...))))...((((.((((((....))))))))))---------)))))))).)))......((((.....))))..... ( -38.10) >DroSim_CAF1 16107 114 + 1 C-----GAGAGUCCCGAGUCCU-UUGUCCUGGUGUUUACAAAAAGGAUGCGCGUGCUGCUGGGGUCGGCAGAUGCCAGGGAUGCCAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUU .-----.......(((.(((((-((((((((((((((.......))))))((((.((((((....)))))))))))))))....)))))))).)))......((((.....))))..... ( -36.20) >DroEre_CAF1 16497 101 + 1 UUGUGUGUGCUG--AGAGUCCU-UUGUCCUGGUGUUUACAAGAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGCC----------------AUGGAAAGCAGAUUGAAAUGAUCGAAUU .......((((.--....(((.-..(((((..((....))...)))))(.((((.((((((....)))))))))))----------------..))).))))((((.....))))..... ( -32.40) >DroYak_CAF1 17206 110 + 1 G-----GGGAGG--AGAGUCCU-UUGUCCUGGUGUUUACAAAAAGGAUGCGCGUGCUGCUGGGGUCGGCAGAUGCCAGGG--GACAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUU .-----......--...(((((-(((((((.((((((.......))))))((((.((((((....))))))))))...))--))))))))))..........((((.....))))..... ( -37.20) >consensus C_____GAGAGUCCAGAGUCCU_UUGUCCUGGUGUUUACAAAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGCC________CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUU ...............(..(((...((((((.((((((.......))))))((((.((((((....))))))))))............)))))).)))...).((((.....))))..... (-27.56 = -28.56 + 1.00)

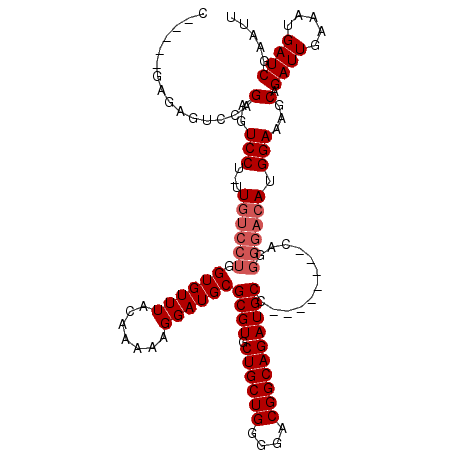
| Location | 1,343,896 – 1,344,005 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1343896 109 + 23771897 AAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGC---------CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAUC-UCGACCGGAGAUC-GAAUCGCAGUGCC ....((.((((((((((((((....))))))...(---------(((....).)))...(((((((((.....)).))))))).))))((((-((.....))))))-....))))...)) ( -40.20) >DroSec_CAF1 16148 109 + 1 AAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGC---------CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAUC-UCGGGCGGAGAUC-GAAUCGCAGCGCC ........((((.((((((((....)))))(((.(---------(((....).)))...(((((((((.....)).))))))).....((((-((.....))))))-..)))))))))). ( -40.40) >DroSim_CAF1 16141 118 + 1 AAAAGGAUGCGCGUGCUGCUGGGGUCGGCAGAUGCCAGGGAUGCCAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAUC-UCGGGCGGAGAUC-GAAUCGCAGUGCC ....((.((((((((((.((((.((((((....)))...))).)))).)).))))....(((((((((.....)).))))))).....((((-((.....))))))-....))))...)) ( -41.60) >DroEre_CAF1 16534 103 + 1 AGAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGCC----------------AUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAAC-UCGGGCGGAGAUCCGAAUCGCAGUACC ....((.((((((..((((((....))))))...((----------------(((....(((((((((.....)).))))))).)))))...-(((((......)))))..))).))))) ( -34.90) >DroYak_CAF1 17238 118 + 1 AAAAGGAUGCGCGUGCUGCUGGGGUCGGCAGAUGCCAGGG--GACAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAUCUUCGUGCGGAGAUCGGCAUCGCAGUGCC ........((((.((((((((....)))))((((((.(..--..)......((((....(((((((((.....)).))))))).))))(((((((....)))))))))))))))))))). ( -45.30) >consensus AAAAGGAUGCGCGUGCUGCUGGGGACGGCAGAUGCC________CAGGGGACAUGGAAAGCAGAUUGAAAUGAUCGAAUUUGCACAUGGAUC_UCGGGCGGAGAUC_GAAUCGCAGUGCC ....((.(((((((.((((((....))))))))))................((((....(((((((((.....)).))))))).))))((((.((....)).))))......)))...)) (-30.30 = -30.70 + 0.40)

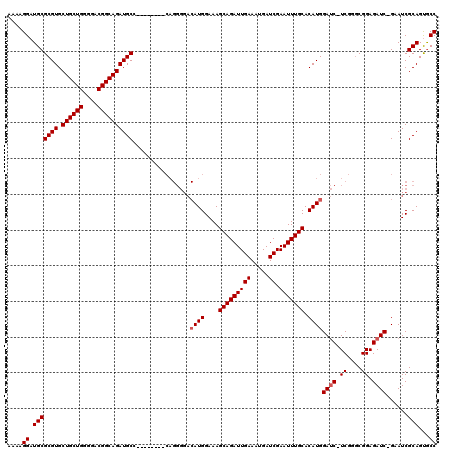
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:53 2006