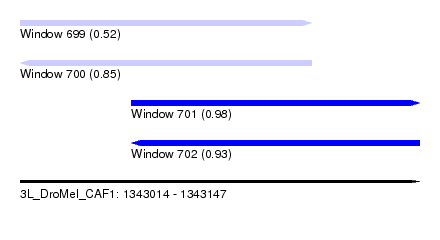
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,343,014 – 1,343,147 |
| Length | 133 |
| Max. P | 0.980620 |

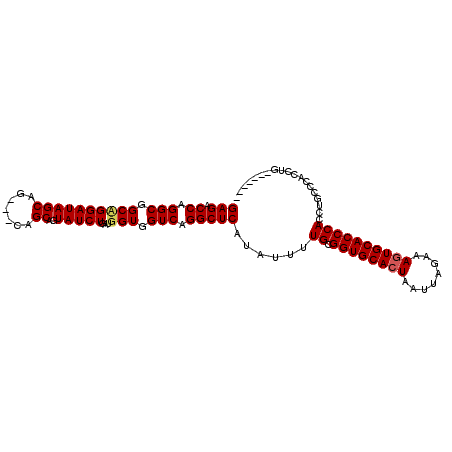
| Location | 1,343,014 – 1,343,111 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -27.51 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
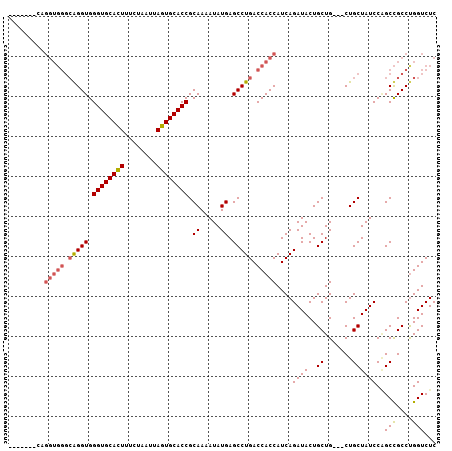
>3L_DroMel_CAF1 1343014 97 + 23771897 GAGACCGGGCGGCUGGAUAGCAG---CAGCAGUAUCUGAUGGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCUGCCCACCUG------- .(((....((.((((.....)))---).))....)))...(((((.((((.........((.((((((((........)))))))))))))).)))))..------- ( -37.40) >DroSim_CAF1 15259 104 + 1 GAGACCAGGCGGCAGGAUAGCAG---CAGCAGUAUCUGAUGGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAAUGCACCCACCCGCCCACCUGCCCACCU ......(((.((((((...((.(---(((.((((((((((....)))))....)))))))))((((((............)))))).....))...))))))..))) ( -32.10) >DroEre_CAF1 15636 91 + 1 GAGACCAGGCAGCCGGAUAGCAGCAGCAGCAGUAUCUGAUGGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCU---------------- (((.((.(((.((((((((.(.((....)).))))))...))).))).)))))......((.((((((((........))))))))))...---------------- ( -30.60) >consensus GAGACCAGGCGGCAGGAUAGCAG___CAGCAGUAUCUGAUGGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCUGCCCACCUG_______ (((.((.(((.((((((((((.......))..)))))...))).))).)))))......((.((((((((........))))))))))................... (-27.51 = -28.07 + 0.56)

| Location | 1,343,014 – 1,343,111 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -25.76 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1343014 97 - 23771897 -------CAGGUGGGCAGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACCAUCAGAUACUGCUG---CUGCUAUCCAGCCGCCCGGUCUC -------..(((((.(((((.((((((((........)))))))).((.....)).))))).)))))...((((...((.(---(((.....)))).))...)))). ( -40.60) >DroSim_CAF1 15259 104 - 1 AGGUGGGCAGGUGGGCGGGUGGGUGCAUUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACCAUCAGAUACUGCUG---CUGCUAUCCUGCCGCCUGGUCUC ((((((.(((((((.(((((.((((((((........)))))))).((.....)).))))).)))))....((((..((..---..)))))).))))))))...... ( -40.30) >DroEre_CAF1 15636 91 - 1 ----------------AGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACCAUCAGAUACUGCUGCUGCUGCUAUCCGGCUGCCUGGUCUC ----------------.((((((((((((........))))))))((.........)).....))))...((((...((.((((........)))).))...)))). ( -29.60) >consensus _______CAGGUGGGCAGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACCAUCAGAUACUGCUG___CUGCUAUCCAGCCGCCUGGUCUC .........(((((.(((((.((((((((........)))))))).((.....)).))))).)))))....((((..((.......))))))..(((....)))... (-25.76 = -27.10 + 1.34)

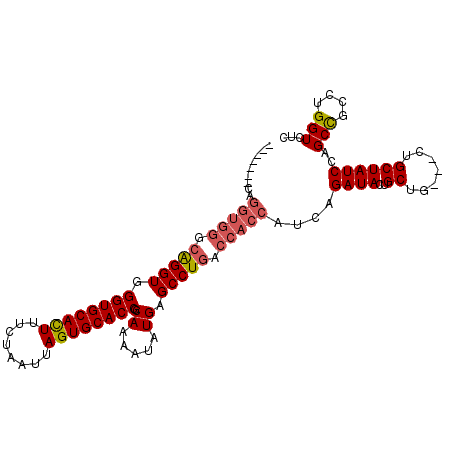
| Location | 1,343,051 – 1,343,147 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
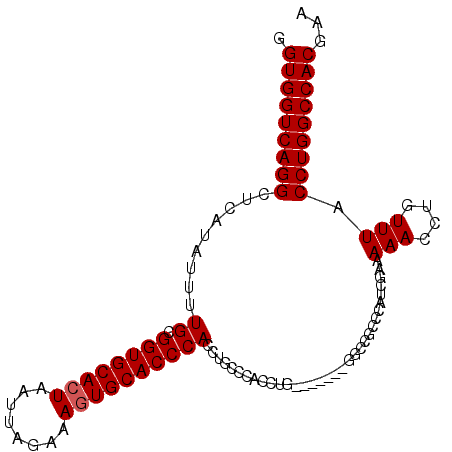
>3L_DroMel_CAF1 1343051 96 + 23771897 GGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCUGCCCACCUG--------ACCACCCAUCGAAAAACCUGUUUACCUGGCCACGAA ((((((((((.........((.((((((((........))))))))))........))))--------))))))..(((.....((.........))...))). ( -34.43) >DroSim_CAF1 15296 104 + 1 GGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAAUGCACCCACCCGCCCACCUGCCCACCUGGCCGCCCAUCGAAAAACCUGUUUACCUGGCCACGAA (((((.((((.........((.((((((............))))))))........)))).)))))((((((.....((.......))......)))))).... ( -29.53) >DroEre_CAF1 15676 88 + 1 GGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCU----------------GGCCGCCCAUCGAAAAACCUGUUUACCUGGCCACCAA ((((((((((.........((.((((((((........))))))))))..(----------------((....)))................)))))))))).. ( -35.20) >consensus GGUGGUCAGGCUCAUAUUUUGCGGUGCACUAAUUAGAAAGUGCACCCACCUGCCCACCUG_______GGCCGCCCAUCGAAAAACCUGUUUACCUGGCCACGAA .(((((((((.........((.((((((((........)))))))))).................................(((....))).)))))))))... (-27.33 = -27.67 + 0.33)

| Location | 1,343,051 – 1,343,147 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -28.01 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
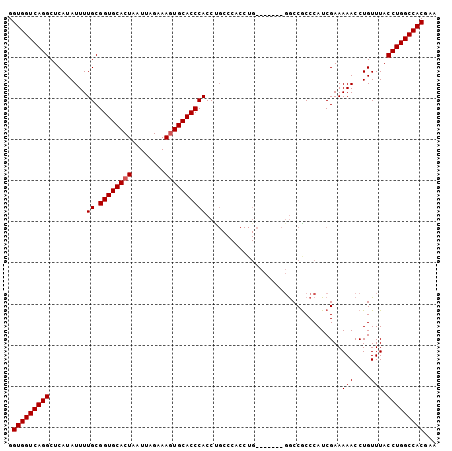
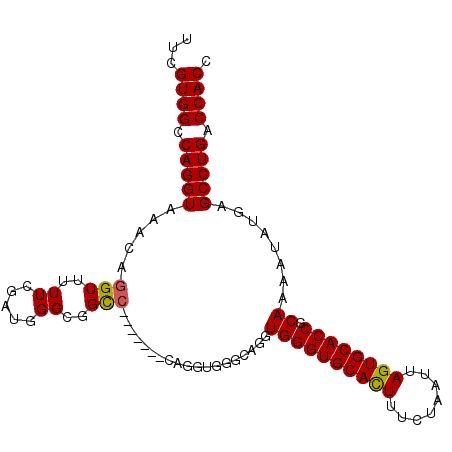
>3L_DroMel_CAF1 1343051 96 - 23771897 UUCGUGGCCAGGUAAACAGGUUUUUCGAUGGGUGGU--------CAGGUGGGCAGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACC .(((.((((.(.....).))))...)))..((((((--------(((((...((..((((((((((........)))))))).)).....)).))))))))))) ( -42.80) >DroSim_CAF1 15296 104 - 1 UUCGUGGCCAGGUAAACAGGUUUUUCGAUGGGCGGCCAGGUGGGCAGGUGGGCGGGUGGGUGCAUUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACC ....(((((.(.....)..((((......)))))))))(((((.(((((.......((((((((((........)))))))).))........))))).))))) ( -39.26) >DroEre_CAF1 15676 88 - 1 UUGGUGGCCAGGUAAACAGGUUUUUCGAUGGGCGGCC----------------AGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACC ..(((((.(((((.....(((..((.....))..)))----------------...((((((((((........)))))))).))........))))).))))) ( -34.70) >consensus UUCGUGGCCAGGUAAACAGGUUUUUCGAUGGGCGGCC_______CAGGUGGGCAGGUGGGUGCACUUUCUAAUUAGUGCACCGCAAAAUAUGAGCCUGACCACC ...((((.(((((.....(((..((.....))..)))...................((((((((((........)))))))).))........))))).)))). (-28.01 = -27.90 + -0.11)

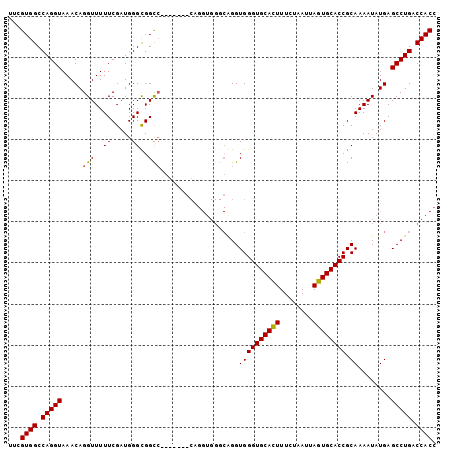
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:51 2006