
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,315,283 – 13,315,415 |
| Length | 132 |
| Max. P | 0.701994 |

| Location | 13,315,283 – 13,315,376 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -26.93 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
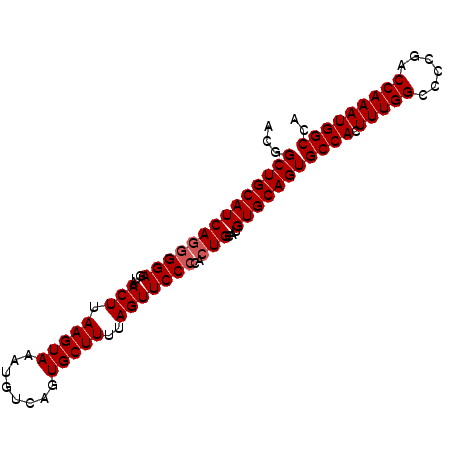
>3L_DroMel_CAF1 13315283 93 + 23771897 ACGGCUGCAUCAUGGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCCACUGCAUGUGCAGUGCCACUUUGGCCCCUACCAAAUGGCCA ...(((((((((((((.(.((((.(((((........)))))..))))))))).))...)))))))((((.(((((......))))))))).. ( -30.20) >DroEre_CAF1 544 93 + 1 ACGGCUGCAUCAGGGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCAACUGCAUGUGCAGUGCCACUUUGGCCCCGGCCAAAUGGCCA ...((((((((((((((...(((.(((((........)))))..)))))))..)))...)))))))((((.((((((....)))))))))).. ( -34.30) >DroYak_CAF1 376 93 + 1 ACGGCUGCAUCAGGGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCGCACUGCAUGUGCAGUGCCACUUUGGCCCCGACCAAAUGGCCA ..((((......((((.(.((((.(((((........)))))..)))).)(((((((....)))))))........))))........)))). ( -31.04) >consensus ACGGCUGCAUCAGGGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCCACUGCAUGUGCAGUGCCACUUUGGCCCCGACCAAAUGGCCA ...((((((((((((((...(((.(((((........)))))..)))))))..)))...)))))))((((.(((((......))))))))).. (-26.93 = -27.60 + 0.67)

| Location | 13,315,296 – 13,315,415 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13315296 119 + 23771897 GGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCCACUGCAUGUGCAGUGCCACUUUGGCCCCUACCAAAUGGCCAAAAAGCCAAAAAC-AAUCCAGACCCGCAAUCUCAGACAUC (((.(.((((.(((((........)))))..))))))))(((((....)))))((((.(((((......)))))))))...............-.......................... ( -26.40) >DroEre_CAF1 557 118 + 1 GGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCAACUGCAUGUGCAGUGCCACUUUGGCCCCGGCCAAAUGGCCAAAAGGCCAAAAACC-AUCCC-UCCCGCAAUCUCAGACAUC (((((.((((.(((((........)))))..))))....(((((....))))).....(((((((..((((....))))....)))))))....-....)-))))............... ( -36.20) >DroYak_CAF1 389 120 + 1 GGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCGCACUGCAUGUGCAGUGCCACUUUGGCCCCGACCAAAUGGCCAAAGAGCCAAAAAACAAUCCCGUCCCGCAAUCUCAGACAUC (((((.((((.(((((........)))))..)))).)(((((((....)))))))..((((((((..........)))))))).............))))(((...........)))... ( -36.30) >consensus GGGAGUAACUUAAGUAAAUGUCAGUGCUUUUAGUUCCCCACUGCAUGUGCAGUGCCACUUUGGCCCCGACCAAAUGGCCAAAAAGCCAAAAACCAAUCCCGUCCCGCAAUCUCAGACAUC ((((...((....))........(.((((((........(((((....)))))((((.(((((......)))))))))..)))))))..............))))............... (-26.47 = -26.37 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:44 2006