
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,302,741 – 13,302,876 |
| Length | 135 |
| Max. P | 0.963259 |

| Location | 13,302,741 – 13,302,837 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -15.24 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13302741 96 - 23771897 UUUUGCAUUUGCAGUUUACUCGGUUAACCGCUUUUCUAGCGGAUUUAACACAUUUC--GGCCUCAGCUGGGC----------GACAACAGAACAGUGGAU-----UGUUUUCG---- ..........((((((((((.(((((((((((.....)))))..))))).....((--(.((......)).)----------))........))))))))-----))).....---- ( -26.10) >DroSim_CAF1 65393 96 - 1 UUUUGCAUUUGCAGUUUACUCGGUUAACCGCUUUUCUAGCGGAUUUAACACAUUUC--GGCCUCAGCUGGGC----------GACAACUGAACAGUGGAU-----UGUUUUUG---- .....((...(((((((((((((((..(((((.....)))))............((--(.((......)).)----------)).))))))...))))))-----)))...))---- ( -28.10) >DroEre_CAF1 65810 96 - 1 UUUUGCAUUUGCAGUUUACUCGGUUAACCGCGUUUCUAGCGGAUUUAACACAUUUC--GGCCUCAGCCGGGC----------GACAACUGAACAGUGGAU-----UGCUCUUA---- ..........(((((((((((((((..((((.......))))...........(((--(((....)))))).----------...))))))...))))))-----))).....---- ( -28.60) >DroWil_CAF1 82539 95 - 1 UUUUGCAUUUGCAGUUUACUCGGUUAACCGCUUUUAUAGCGGAUUUAACACAUUUUCAGGCCU----UGGGCUCAUAACAUUGACCAAUGCACAGUGGC------------------ .....(((((((((......)(((((((((((.....)))))................((((.----..)))).......))))))..)))).))))..------------------ ( -21.70) >DroYak_CAF1 70819 96 - 1 UUUUGCAUUUGCAGUUUACUCGGUUAACCGCUUUUCUAGCGGAUUUAACACAUUUC--GGCCUCAGCUGGGC----------GACAACUGAACAGUGGAU-----UGUUUUUG---- .....((...(((((((((((((((..(((((.....)))))............((--(.((......)).)----------)).))))))...))))))-----)))...))---- ( -28.10) >DroAna_CAF1 61745 97 - 1 UUUUGCAUUU-CAGUUUACUCGGUUAACCGCUUUUAUAGCGGAUUUAGCACAUUUC--AGCCUUGGC----C----------AAUA---CCGUGGUGGAUGUACUGGCUUUUGUGGA .((..((..(-((((.......((((((((((.....)))))..)))))(((((..--.(((.(((.----.----------....---))).))).))))))))))....))..)) ( -25.80) >consensus UUUUGCAUUUGCAGUUUACUCGGUUAACCGCUUUUCUAGCGGAUUUAACACAUUUC__GGCCUCAGCUGGGC__________GACAACUGAACAGUGGAU_____UGUUUUUG____ ...(((....)))(((((((..((((((((((.....)))))..)))))..........((((.....)))).....................)))))))................. (-15.24 = -16.13 + 0.89)



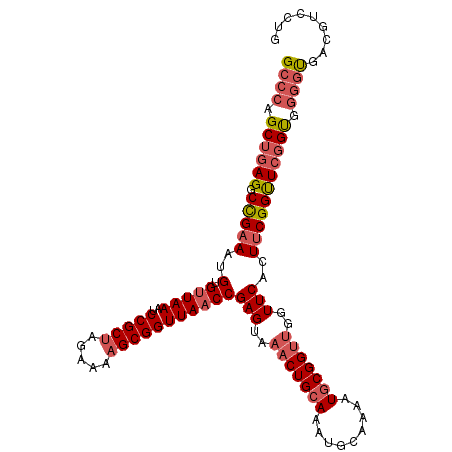
| Location | 13,302,767 – 13,302,876 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -33.52 |
| Energy contribution | -34.80 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13302767 109 + 23771897 GCCCAGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAAAGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUUCGGUUCGGUGGGGUGACGUCCUG ((((.((((((.(((((..(.(((((..(((((.....)))))))))))(((..(((((((.........)))))))..)))..))))))))))).))))......... ( -39.90) >DroSec_CAF1 64793 109 + 1 GCCCAGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAAAGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUUCGGCUCGGUGGGGUGAUGUCCUG ((((.((((((.(((((..(.(((((..(((((.....)))))))))))(((..(((((((.........)))))))..)))..))))))))))).))))......... ( -42.30) >DroSim_CAF1 65419 109 + 1 GCCCAGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAAAGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUUCGGUUCGGUGGGGUGAUGUCCUG ((((.((((((.(((((..(.(((((..(((((.....)))))))))))(((..(((((((.........)))))))..)))..))))))))))).))))......... ( -39.90) >DroEre_CAF1 65836 109 + 1 GCCCGGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAACGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUUCGGUUCGGUGGGGUGACAUCCUG ((((.((((((.(((((..(.(((((..((((.......))))))))))(((..(((((((.........)))))))..)))..))))))))))).))))......... ( -40.50) >DroYak_CAF1 70845 109 + 1 GCCCAGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAAAGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUCCGGUUCGGUGGGGUGACGUCCUG ((((.((((((.(((....(.(((((..(((((.....)))))))))))(((..(((((((.........)))))))..)))....))))))))).))))......... ( -37.60) >DroAna_CAF1 61777 100 + 1 G----GCCAAGGCUGAAAUGUGCUAAAUCCGCUAUAAAAGCGGUUAACCGAGUAAACUG-AAAUGCAAAAUGCGG----UUCAUUUCGGUUCUGCUGGGCUCUUGCCCG (----((.(..(((((((((.(((....(((((.....)))))................-....((.....))))----).)))))))))..))))((((....)))). ( -33.20) >consensus GCCCAGCUGAGGCCGAAAUGUGUUAAAUCCGCUAGAAAAGCGGUUAACCGAGUAAACUGCAAAUGCAAAAUGCGGUUGGUUCACUUCGGUUCGGUGGGGUGACGUCCUG ((((.((((((.(((((..(.(((((..(((((.....)))))))))))(((..(((((((.........)))))))..)))..))))))))))).))))......... (-33.52 = -34.80 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:37 2006