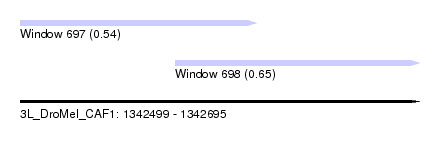
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,342,499 – 1,342,695 |
| Length | 196 |
| Max. P | 0.650772 |

| Location | 1,342,499 – 1,342,615 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -24.56 |
| Energy contribution | -25.72 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1342499 116 + 23771897 ACAGGAGCAGGAGAUUCUGUGGUUGCUUGCCCCUGCAAGGAAGCAUUUGCGCGUGUCUCCU----GGCUCCCCUCCUUUGGCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCAUCAAAU ...((((((((((((.(((..(.(((((.((.......))))))).)..)).).)))))))----.)))))((......))..........((((.((.........)).))))...... ( -35.70) >DroSec_CAF1 14751 116 + 1 ACAGGAGCUGGAGAUUCUGUGGUUGCUUGCUCCUUCAAGGGAGCAGUUGCGCGUGUCUACU----GGCUCCCCUCCUUUGGCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAU ..((((((..((((....((((((((((((((((....))))))))..))..(.(((....----))).).........))))))..))))..).)))))......((((...))))... ( -38.30) >DroSim_CAF1 14761 116 + 1 ACAGGAGCUGGAGAUUCUGUGGUUGCUUGCUCCUCCAAGGGAGCAGUUGCGCGUGUCUCCU----GGCUCCCCUCCUUUGGCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAU ...((((((((((((..((((...(((.((((((....)))))))))..)))).)))))).----)))))).....((((((..........(((.((.........)).))))))))). ( -39.60) >DroEre_CAF1 15081 111 + 1 ACAGAAGCAGGAGAUUCUGUGGUUGCUUGCCCCUGC-----GGCAGUUGCACGUGCCUCCU----GACUCCCUUCCCUUGGCCACUUUCUCUUGGCUCCUUUCUUCUGACCCCGUCAAAU .((((((((((((...((((..(((((.((....))-----)))))..))).)...)))))----).............(((((........))))).....))))))............ ( -32.90) >DroYak_CAF1 15831 115 + 1 ACAGGAGCUGGAGAUCCUGCGGUUGCUUGCCCCUGC-----GGCAGUUGCACGUGCCUCCUAGCUGCCUCCCUUCCCCUGGCCACUUUCUCUUGGCUCCUUUCUUUUGACCCCAUCAAAU ..((((((((((((....(.((((((........))-----(((((((((....)).....)))))))...........)))).)..)))).))))))))....(((((.....))))). ( -33.60) >consensus ACAGGAGCUGGAGAUUCUGUGGUUGCUUGCCCCUGCAAGG_AGCAGUUGCGCGUGUCUCCU____GGCUCCCCUCCUUUGGCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAU ..((((((((((((....((((((((.(((..((((......))))..))).))(((........)))...........))))))..)))))).))))))......(((.....)))... (-24.56 = -25.72 + 1.16)

| Location | 1,342,575 – 1,342,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1342575 120 + 23771897 GCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCAUCAAAUUGACUGCUGCCAAAUGUUUUCCUCGCUGCGCAUUAAGCACACAGUGCUUCAUUUGGCCAAGUACAGUGGAGCAGGGAUAU .......(((((..(((((.......(((.....))).(((((((...((((((((.......(((((.((.....))...)))))...))))))))..))).))))))))))))))... ( -32.60) >DroSec_CAF1 14827 120 + 1 GCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAUUGACUGCUGCCAAAUGUUUUCCUCGCUGCGCAUUAAGCACACAGUGCUUCAUUUGGCCAAGUACCGUGCAGCAGGGCUAA ((...........(((..(..((...((((...))))....))..)..))).........(((.(((((((...((((((...)))))).....((.......))))))))))))))... ( -31.50) >DroSim_CAF1 14837 120 + 1 GCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAUUGACUGCUGCCAAAUGUUUUCCUCGCUGCGCAUUAAGCACACAGUGCUUCAUUUGGCCAAGUACAGUGCAGCAGGGCUAA ((...........(((..(..((...((((...))))....))..)..))).........(((.(((((((...((((((...))))))....((........))))))))))))))... ( -30.60) >DroEre_CAF1 15152 120 + 1 GCCACUUUCUCUUGGCUCCUUUCUUCUGACCCCGUCAAAUUGACUGCUGCCAAAUGUUUUCAACGCUGCGCAUUAAGCGCACAGUGCUUCAUUUGGCCAAGUACAGUGCGGCGGGGCUAA ((((........)))).............(((((((..(((((((...((((((((......((..(((((.....)))))..))....))))))))..))).))))..))))))).... ( -44.70) >DroYak_CAF1 15906 120 + 1 GCCACUUUCUCUUGGCUCCUUUCUUUUGACCCCAUCAAAUUGACUGCUGCCAAAUGUUUUCCUCGCUGCGCAUUAAGCACACAGUGCUUCAUUUGGCCAAGUACAGUGCAGCAGGGCUAA (((........(((((..(..((.(((((.....)))))..))..)..)))))...........(((((((...((((((...))))))....((........)))))))))..)))... ( -33.50) >consensus GCUACUUUCUCUGGGCUCCUUUCUUCUGACCCCGUCAAAUUGACUGCUGCCAAAUGUUUUCCUCGCUGCGCAUUAAGCACACAGUGCUUCAUUUGGCCAAGUACAGUGCAGCAGGGCUAA (((..........))).............(((.((...(((((((...((((((((.......(((((.((.....))...)))))...))))))))..))).))))...)).))).... (-26.82 = -27.18 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:48 2006