
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,279,794 – 13,279,905 |
| Length | 111 |
| Max. P | 0.648384 |

| Location | 13,279,794 – 13,279,905 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.05 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13279794 111 - 23771897 GG-U----GGCCUAUUGGGCUAUUGGUUGCCAUGUCAACGGGGCGGAAAUCAGGCAACAUCCGCUCCAUAAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUGCAG--U--UUCAGA ((-(----((((.....)))))))..(((((((((.(((((((((((.....(....).))))))))((((((((....)).))))))......))).))).)).))))--.--...... ( -35.80) >DroVir_CAF1 33922 96 - 1 GA-------G-------------CGUCUG--CUGUCAACAAACCGGAGAUCAGGCAACAUCCGUUCCAUAAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUGCUU--CCAUUCAGA .(-------(-------------(((..(--((((.(((((((((((.....(....)(((((((.....))))))).))))))))))).))).))..)))))......--......... ( -37.70) >DroPse_CAF1 45391 88 - 1 GGAU----GG---------------------CUGUCAAUGGGUCGGAAAUCAGGCAACAUCCGCUCCAUGAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUACUG--C--GUC--- ((((----((---------------------(.....((((..((((.....(....).))))..))))((((((((.....)))))))))).)))))((((......)--)--)).--- ( -28.60) >DroGri_CAF1 38019 96 - 1 GA-------G-------------CGGCUG--CUGUCAACAAACCGGAGAUCAGGCAACAUCCGUUUCAUAAACGGAUGUCCGGUUUGUUCGCACGUUCACGCUGUGCUG--CCAUUGAGA (.-------(-------------((((.(--((((.(((((((((((.....(....)(((((((.....))))))).))))))))))).))).(....)))...))))--))....... ( -37.00) >DroAna_CAF1 41909 117 - 1 GG-UUGGCUGCCCAUUGGGAUAUUGGUCGGCCUGUCAACGGGGCGGAAAUCAGGCAACAUCCGCUCCAUAAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUGCUGUGU--UUCAGG .(-(((((.(((.((..(....)..)).)))..))))))((((((((.....(....).))))))))((((((((....)).)))))).((((((....)).))))(((...--..))). ( -39.70) >DroPer_CAF1 45450 87 - 1 GG-U----GG---------------------CUGUCAAUGGGUCGGAAAUCAGGCAACAUCCGCUCCAUGAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUACUG--C--GUC--- .(-(----((---------------------((((.((..(..((((.....(....)(((((.((...)).))))).))))..)..)).))).).))))((......)--)--...--- ( -25.50) >consensus GG_U____GG______________GG_UG__CUGUCAACGGGCCGGAAAUCAGGCAACAUCCGCUCCAUAAACGGAUAUCCGGUUUGUUCGCACGUUCACGCUGUGCUG__C__UUCAGA ................................(((.(((((((((((.....(....)(((((.........))))).))))))))))).)))........................... (-20.71 = -20.05 + -0.66)

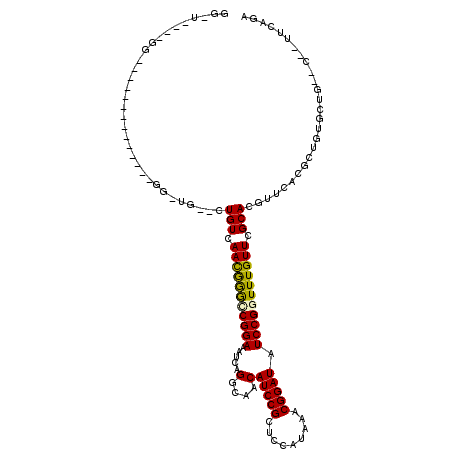
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:23 2006