
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,254,839 – 13,254,941 |
| Length | 102 |
| Max. P | 0.595785 |

| Location | 13,254,839 – 13,254,941 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
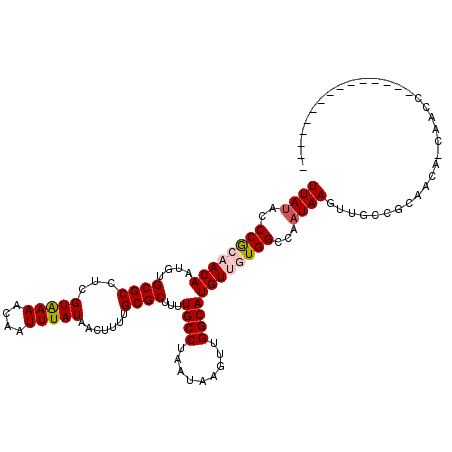
>3L_DroMel_CAF1 13254839 102 - 23771897 UUACGGCCGCAACAAUGUGCGCCUUGUGAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGCGGCAACAGCAGC---------------- ....((((((((((....((((.(((((((....))))))).....))))...((((.........)))))))))))))).....((((((....).)))))---------------- ( -37.50) >DroVir_CAF1 6785 117 - 1 UUAUACCCGCAACAAUGUGCGCCUCGUAAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGCCGCAUAA-UAACCCCCUCCAGCCCACGG ......((((((((....((((...(((((....))))).......))))...((((.........))))))))))))...................-.................... ( -20.30) >DroPse_CAF1 19663 108 - 1 UUAUGGCCAACACAAUGUGCGCCUUGUAAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGUGGCAACAACAACCC----------AAGA ...(((((...(((((((((...(((.....)))....(((((((((.((....)).))).))))))))))))))))))))....((((((....).)))))..----------.... ( -30.20) >DroGri_CAF1 6995 111 - 1 UUAUACCCGCAACAAUGUGCGCCUCGUAAAACAAUUUAUAACUUUUGCGCUCUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGCCGCAGAA-UAGCC--CUCCAACCU---- ......((((((((..(.((((...(((((....))))).......)))).).((((.........)))))))))))).......((((..((....-..)).--...))))..---- ( -23.30) >DroMoj_CAF1 11636 93 - 1 UUAUACCCGCAACAAUGUGCGCCUCGUAAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGCCGC------------------------- ......((((((((....((((...(((((....))))).......))))...((((.........))))))))))))...............------------------------- ( -20.30) >DroPer_CAF1 18597 108 - 1 UUAUGGCCAACACAAUGUGCGCCUUGUAAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGUGGCAACAACAACCC----------AAGA ...(((((...(((((((((...(((.....)))....(((((((((.((....)).))).))))))))))))))))))))....((((((....).)))))..----------.... ( -30.20) >consensus UUAUACCCGCAACAAUGUGCGCCUCGUAAAACAAUUUAUAACUUUUGCGCUUUUGCCUAAUAAGUUGGCAUGUUGUGGCCAAUAAGUUGCCGCAACA_CAACC_______________ ((((..((((((((....((((...(((((....))))).......))))...((((.........))))))))))))...))))................................. (-18.48 = -18.95 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:19 2006