
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,252,122 – 13,252,224 |
| Length | 102 |
| Max. P | 0.999165 |

| Location | 13,252,122 – 13,252,224 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13252122 102 + 23771897 CGUUUCUCACUUUUGGAGAGAAUUUUGGGAAAUACAGUAC--------ACAAAGGG------CCUAUAUUCACUUUCAUUUAUUUUCUUUAAAAGUGAAAAUCGCAGUAAAUCUGU ......(((((((((((((((((..((..(((((.((..(--------......).------.)).))))...)..))...))))))))))))))))).....((((.....)))) ( -25.10) >DroSec_CAF1 14878 91 + 1 UGGUUCUCACUUUUGGAGAGAGUUUUGGGAAGUAUGAUAU-------------------------AUAUGCACUUUGAUUUAUUUUCUUCAAAAGUGAAAGUCGCAGUAAAUCUGU .(..(.(((((((((((((((((.(..((..(((((....-------------------------.)))))..))..)...))))))))))))))))).)..)((((.....)))) ( -27.90) >DroSim_CAF1 15362 101 + 1 CGGUUCUCACUUUUGGAGAGAGUUUUGGGAAGUAAGAUAU--------ACAAAGGA-------CUAUAUGCACUUUCAUUUAUUUUCUUCAAAAGUGAAAAUCGCAGUAAAUCUGU .((((.(((((((((((((((((....((((((...((((--------(.......-------.)))))..))))))....))))))))))))))))).))))((((.....)))) ( -33.80) >DroEre_CAF1 14168 102 + 1 CGGUUCUCACUUUUGGAGAGAGUUUUGGGUAGUAAGCUAA--------ACAUAAGG------AACCUAUUCGCUUUCAUUUAUUUUACUCAAAAGUGAAAACCGUAGCCAACCUGU (((((.((((((((((.((((((..(((((..........--------........------.)))))...))))))...........)))))))))).)))))............ ( -24.21) >DroYak_CAF1 15090 116 + 1 CGGUUCUCACUUUUGGAGAGAGUUUUCAGCAGUAAGUUAAUAAAAUAAAAAAAAGGGAUGCAAACUUAUUCGCUUUCAUUUACUUUAUUCAAAAGUGAAAAUCGUAGCCAACCUGC (((((.(((((((((((.(((((....(((((((((((........................)))))))).))).......))))).))))))))))).)))))............ ( -27.46) >consensus CGGUUCUCACUUUUGGAGAGAGUUUUGGGAAGUAAGAUAA________ACAAAAGG_______ACAUAUUCACUUUCAUUUAUUUUCUUCAAAAGUGAAAAUCGCAGUAAAUCUGU .((((.(((((((((((((((((....((((((......................................))))))....))))))))))))))))).))))((((.....)))) (-22.88 = -23.28 + 0.40)


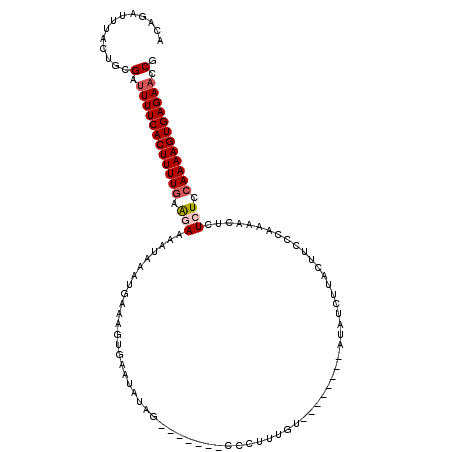
| Location | 13,252,122 – 13,252,224 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -9.50 |
| Energy contribution | -10.14 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13252122 102 - 23771897 ACAGAUUUACUGCGAUUUUCACUUUUAAAGAAAAUAAAUGAAAGUGAAUAUAGG------CCCUUUGU--------GUACUGUAUUUCCCAAAAUUCUCUCCAAAAGUGAGAAACG .(((.....)))((.(((((((((((..(((.(((...((...(..(((((((.------.(......--------)..)))))))..)))..))).)))..))))))))))).)) ( -19.00) >DroSec_CAF1 14878 91 - 1 ACAGAUUUACUGCGACUUUCACUUUUGAAGAAAAUAAAUCAAAGUGCAUAU-------------------------AUAUCAUACUUCCCAAAACUCUCUCCAAAAGUGAGAACCA .(((.....)))....(((((((((((.(((..........(((((.....-------------------------......)))))..........))).))))))))))).... ( -14.95) >DroSim_CAF1 15362 101 - 1 ACAGAUUUACUGCGAUUUUCACUUUUGAAGAAAAUAAAUGAAAGUGCAUAUAG-------UCCUUUGU--------AUAUCUUACUUCCCAAAACUCUCUCCAAAAGUGAGAACCG .(((.....)))((.((((((((((((.(((.........((((..(.....)-------..))))((--------(.....)))............))).)))))))))))).)) ( -20.20) >DroEre_CAF1 14168 102 - 1 ACAGGUUGGCUACGGUUUUCACUUUUGAGUAAAAUAAAUGAAAGCGAAUAGGUU------CCUUAUGU--------UUAGCUUACUACCCAAAACUCUCUCCAAAAGUGAGAACCG ............((((((((((((((((((((..((((((.(((.(((....))------)))).)))--------)))..)))))...............))))))))))))))) ( -27.36) >DroYak_CAF1 15090 116 - 1 GCAGGUUGGCUACGAUUUUCACUUUUGAAUAAAGUAAAUGAAAGCGAAUAAGUUUGCAUCCCUUUUUUUUAUUUUAUUAACUUACUGCUGAAAACUCUCUCCAAAAGUGAGAACCG ((......))..((.(((((((((((((((((((((((.(((((.((...........)).))))).)))))))))))...........((......))..)))))))))))).)) ( -24.20) >consensus ACAGAUUUACUGCGAUUUUCACUUUUGAAGAAAAUAAAUGAAAGUGAAUAUAG_______CCCUUUGU________AUAUCUUACUUCCCAAAACUCUCUCCAAAAGUGAGAACCG .............(.((((((((((((.(((..................................................................))).)))))))))))).). ( -9.50 = -10.14 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:17 2006