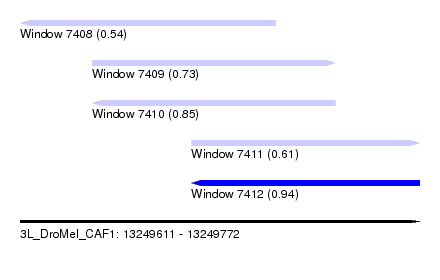
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,249,611 – 13,249,772 |
| Length | 161 |
| Max. P | 0.935790 |

| Location | 13,249,611 – 13,249,714 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13249611 103 - 23771897 GGCACAGGCACAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCACUG---UCUUUUG-----GCU---C (((.(((((...(((------....)))....))))))))(((((((.((((..(((((..(((....))).)))))..))))............)))---))))...-----...---. ( -35.62) >DroSec_CAF1 12388 93 - 1 ----------CAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCACUG---UCUUUUG-----GCU---C ----------(((((------...........)))))((((((((((.((((..(((((..(((....))).)))))..))))............)))---)))..))-----)).---. ( -31.42) >DroSim_CAF1 12890 93 - 1 ----------CAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCACUG---UCUUUUG-----GCU---C ----------(((((------...........)))))((((((((((.((((..(((((..(((....))).)))))..))))............)))---)))..))-----)).---. ( -31.42) >DroEre_CAF1 11659 104 - 1 ----------CAGGCAAAGGCUAAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCGCUG---UCUUUUGGCUUGGCU---U ----------(((((...(((((.(((.....))))))))(((((((.(((((((((((..(((....))).))))).))..........)))).)))---))))...)))))...---. ( -39.50) >DroYak_CAF1 12585 99 - 1 ----------CAGGCCCAUGCACAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCACUG---UCUUUUG-----GCU---U ----------.(((((...((.(((((.....))))))).(((((((.((((..(((((..(((....))).)))))..))))............)))---))))..)-----)))---) ( -35.02) >DroAna_CAF1 4426 96 - 1 ----------CAGG---------AGGCCCAGGGUCUGGCCAAGGAGAUUGGCGAACGACAUGGUAAUUGCCAGUCGUCCGCUCAUUACCCACUCACUGGCUUCUUUUG-----GCUGGCU ----------.((.---------.((((.((((...(((((..(((..(((((.(((((.((((....))))))))).))).)).......)))..))))).)))).)-----)))..)) ( -36.30) >consensus __________CAGGC______AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUGCUCAUUACACGCUCACUG___UCUUUUG_____GCU___C ..........(((...........(((((......)))))..((((((.((...(((((..(((....))).))))))).)))))).........)))...................... (-22.98 = -22.78 + -0.19)


| Location | 13,249,640 – 13,249,738 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13249640 98 + 23771897 CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUCU------GCCUGUGCCUGUGCCUGUGCCUGGUUUUGGCCACAUUGC ..(((((((((......)).)))))))((((..(((....((((((((...(((.(.(.------....).).))).))))).))))))....))))....... ( -32.30) >DroSec_CAF1 12417 86 + 1 CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUCU------GCCUG------------UGCCUGGUUUUGGCCACAUUGC ..(((((((((......)).)))))))((((..(((....((((((((...........------)))))------------.))))))....))))....... ( -30.30) >DroSim_CAF1 12919 86 + 1 CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUCU------GCCUG------------UGCCUGGUUUUGGCCACAUUGC ..(((((((((......)).)))))))((((..(((....((((((((...........------)))))------------.))))))....))))....... ( -30.30) >DroEre_CAF1 11693 92 + 1 CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUUAGCCUUUGCCUG------------UGCCUGGUUUUGGCCACAUUAC ..(((((((((......)).)))))))((((..(((....((((((((.....((....))....)))))------------.))))))....))))....... ( -31.50) >DroYak_CAF1 12614 92 + 1 CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUGUGCAUGGGCCUG------------UGCCUGGUUUUGGCCACAUUAC ..(((((((((......)).)))))))((((..(((....((((((((((...((....))..)))))))------------.))))))....))))....... ( -33.80) >DroAna_CAF1 4461 83 + 1 CGGACGACUGGCAAUUACCAUGUCGUUCGCCAAUCUCCUUGGCCAGACCCUGGGCCU---------CCUG------------CUCCUAGUCCUGGCCACGGUGC (((((((((((......))).)))))))).....(.((.(((((((..(..((((..---------...)------------).))..)..))))))).)).). ( -34.60) >consensus CAGACGACGGGCAAUUACCAUGUCGUUGGCUCCCCAUCUUGGCCAGGCUUUUAGCCUCU______GCCUG____________UGCCUGGUUUUGGCCACAUUGC ..(((((((((......)).)))))))((((.........((((((((...................................))))))))..))))....... (-21.24 = -21.80 + 0.56)

| Location | 13,249,640 – 13,249,738 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.04 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13249640 98 - 23771897 GCAAUGUGGCCAAAACCAGGCACAGGCACAGGCACAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ((..((((.((.......)))))).)).(((((...(((------..((((.....)))).)))..((((((.(((((......))))...).))))))))))) ( -38.80) >DroSec_CAF1 12417 86 - 1 GCAAUGUGGCCAAAACCAGGCA------------CAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ((......))......(((((.------------..(((------..((((.....)))).)))..((((((.(((((......))))...).))))))))))) ( -31.30) >DroSim_CAF1 12919 86 - 1 GCAAUGUGGCCAAAACCAGGCA------------CAGGC------AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ((......))......(((((.------------..(((------..((((.....)))).)))..((((((.(((((......))))...).))))))))))) ( -31.30) >DroEre_CAF1 11693 92 - 1 GUAAUGUGGCCAAAACCAGGCA------------CAGGCAAAGGCUAAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ........(((.......))).------------(((((...(((((.(((.....))))))))..((((((.(((((......))))...).))))))))))) ( -36.20) >DroYak_CAF1 12614 92 - 1 GUAAUGUGGCCAAAACCAGGCA------------CAGGCCCAUGCACAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ........(((.......))).------------(((((....((.(((((.....)))))))...((((((.(((((......))))...).))))))))))) ( -33.20) >DroAna_CAF1 4461 83 - 1 GCACCGUGGCCAGGACUAGGAG------------CAGG---------AGGCCCAGGGUCUGGCCAAGGAGAUUGGCGAACGACAUGGUAAUUGCCAGUCGUCCG ......(((((((..((.((.(------------(...---------..)))).))..))))))).((((((((((((((......))..))))))))).))). ( -37.70) >consensus GCAAUGUGGCCAAAACCAGGCA____________CAGGC______AGAGGCUAAAAGCCUGGCCAAGAUGGGGAGCCAACGACAUGGUAAUUGCCCGUCGUCUG ......(((((....((...................)).........(((((...)))))))))).((((((.(((((......))))...).))))))..... (-23.04 = -23.04 + 0.00)



| Location | 13,249,680 – 13,249,772 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13249680 92 + 23771897 GGCCAGGCUUUUAGCCUCU------GCCUGUGCCUGUGCCUGUGCCUGGUUUUGGCCACAUUGCCAGUG------GCCCUGGGAUUGGCCAACUGCCCGAAUGG ((((((((...........------))))).)))((.(((....((..(....((((((.......)))------))))..))...)))))............. ( -35.30) >DroSec_CAF1 12457 80 + 1 GGCCAGGCUUUUAGCCUCU------GCCUG------------UGCCUGGUUUUGGCCACAUUGCCAGUG------GCCCUGGGAUUGGCCAACUGCCCGAAUGG ((((((((...........------)))))------------.))).(((.(((((((...(.((((..------...)))).).)))))))..)))....... ( -33.10) >DroSim_CAF1 12959 80 + 1 GGCCAGGCUUUUAGCCUCU------GCCUG------------UGCCUGGUUUUGGCCACAUUGCCAGUG------GCCCUGGGUUUGGCCAACUGCCCGAAUGG ((((((((...........------)))))------------.))).(((.(((((((.((..((((..------...)))))).)))))))..)))....... ( -31.90) >DroEre_CAF1 11733 86 + 1 GGCCAGGCUUUUAGCCUUAGCCUUUGCCUG------------UGCCUGGUUUUGGCCACAUUACCAGUG------GCCCUGGGAUUGGCCAACUGCCCGAAUGG ((((((((.....((....))....)))))------------.))).(((.(((((((.(((.((((..------...))))))))))))))..)))....... ( -33.90) >DroYak_CAF1 12654 86 + 1 GGCCAGGCUUUUAGCCUGUGCAUGGGCCUG------------UGCCUGGUUUUGGCCACAUUACCAGUG------GCCCUGGGAUUGGCCAACUGCCCCAAUGG .(((((((.....))))).))..((((...------------((.(..(((((((((((.......)))------)))..)))))..).))...))))...... ( -36.30) >DroAna_CAF1 4501 83 + 1 GGCCAGACCCUGGGCCU---------CCUG------------CUCCUAGUCCUGGCCACGGUGCACGUGCCCGGUGCCCGGGGACUGGCCAACUGCCCCAAUGG ((((((.((((((((..---------.(((------------....)))..((((.((((.....)))).)))).)))))))).)))))).............. ( -40.80) >consensus GGCCAGGCUUUUAGCCUCU______GCCUG____________UGCCUGGUUUUGGCCACAUUGCCAGUG______GCCCUGGGAUUGGCCAACUGCCCGAAUGG ((((((((...................................))))))))(((((((.(((.((((...........))))))))))))))............ (-21.17 = -21.65 + 0.48)



| Location | 13,249,680 – 13,249,772 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -35.29 |
| Consensus MFE | -24.41 |
| Energy contribution | -23.75 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13249680 92 - 23771897 CCAUUCGGGCAGUUGGCCAAUCCCAGGGC------CACUGGCAAUGUGGCCAAAACCAGGCACAGGCACAGGCACAGGC------AGAGGCUAAAAGCCUGGCC .(((...(.((((.((((........)))------))))).)...)))(((.......)))...(((.(((((...(((------....)))....)))))))) ( -36.00) >DroSec_CAF1 12457 80 - 1 CCAUUCGGGCAGUUGGCCAAUCCCAGGGC------CACUGGCAAUGUGGCCAAAACCAGGCA------------CAGGC------AGAGGCUAAAAGCCUGGCC ......((....(((((((...((((...------..)))).....)))))))..)).(((.------------(((((------...........)))))))) ( -32.50) >DroSim_CAF1 12959 80 - 1 CCAUUCGGGCAGUUGGCCAAACCCAGGGC------CACUGGCAAUGUGGCCAAAACCAGGCA------------CAGGC------AGAGGCUAAAAGCCUGGCC ......((....(((((((...((((...------..)))).....)))))))..)).(((.------------(((((------...........)))))))) ( -32.50) >DroEre_CAF1 11733 86 - 1 CCAUUCGGGCAGUUGGCCAAUCCCAGGGC------CACUGGUAAUGUGGCCAAAACCAGGCA------------CAGGCAAAGGCUAAGGCUAAAAGCCUGGCC ......((....(((((((...((((...------..)))).....)))))))..)).(((.------------(((((...(((....)))....)))))))) ( -35.90) >DroYak_CAF1 12654 86 - 1 CCAUUGGGGCAGUUGGCCAAUCCCAGGGC------CACUGGUAAUGUGGCCAAAACCAGGCA------------CAGGCCCAUGCACAGGCUAAAAGCCUGGCC ......((((...(((((........(((------(((.......)))))).......))).------------)).))))..((.(((((.....))))))). ( -36.46) >DroAna_CAF1 4501 83 - 1 CCAUUGGGGCAGUUGGCCAGUCCCCGGGCACCGGGCACGUGCACCGUGGCCAGGACUAGGAG------------CAGG---------AGGCCCAGGGUCUGGCC ((....))......((((((.(((.((((.((((.((((.....)))).)).)).((.....------------.)).---------..)))).))).)))))) ( -38.40) >consensus CCAUUCGGGCAGUUGGCCAAUCCCAGGGC______CACUGGCAAUGUGGCCAAAACCAGGCA____________CAGGC______AGAGGCUAAAAGCCUGGCC .......(((..(((((((...((((...........)))).....)))))))..................................(((((...))))).))) (-24.41 = -23.75 + -0.66)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:11 2006