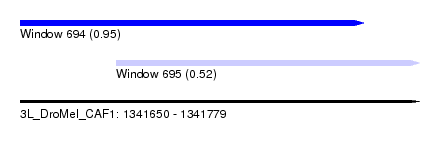
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,341,650 – 1,341,779 |
| Length | 129 |
| Max. P | 0.951431 |

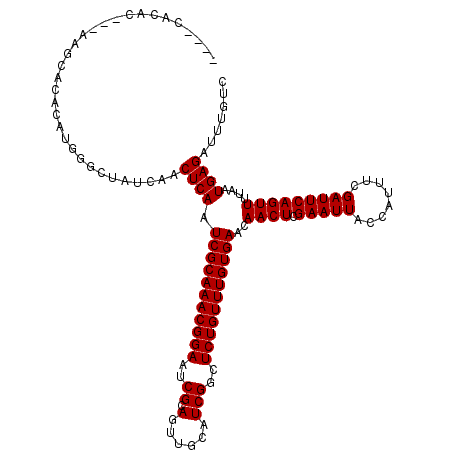
| Location | 1,341,650 – 1,341,761 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1341650 111 + 23771897 ------CAC---AAGCACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ------.((---((((........))).....((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....))))...))). ( -26.30) >DroSec_CAF1 13844 113 + 1 ----CACAC---UAGCACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ----.....---((((........))))....((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....))))....... ( -27.20) >DroSim_CAF1 13894 116 + 1 ----CACACUACUAGCACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ----........((((........))))....((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....))))....... ( -27.20) >DroEre_CAF1 14230 114 + 1 ---ACAGAC---AAACACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ---...(((---(((......(((....))).((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....)))).)))))) ( -28.30) >DroYak_CAF1 14934 117 + 1 CACACAGAC---AAACACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ......(((---(((......(((....))).((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....)))).)))))) ( -28.30) >consensus ____CACAC___AAGCACACAUGGGCUAUCAACUCAAUCGCAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUC ................................((((.(((((((((((..((.(......)))..)))))))))))..((((.(((((........)))))))))....))))....... (-24.40 = -24.40 + 0.00)

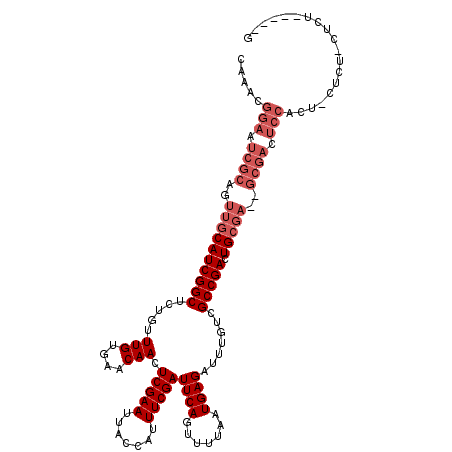
| Location | 1,341,681 – 1,341,779 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -22.78 |
| Energy contribution | -24.78 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1341681 98 + 23771897 CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUG----------------------CUCUGCCCUG .....((....((((..((((((((.....(((....))).(((((.......)))))((((.......)))).......))))).))----------------------).)))))).. ( -23.80) >DroSec_CAF1 13877 113 + 1 CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUGCGA--GCGACUCCACUGCUCUGCUCU-----G .....(((.((((..((((((((((.....(((....))).(((((.......)))))((((.......)))).......))))).)))))--)))).))).............-----. ( -29.80) >DroSim_CAF1 13930 118 + 1 CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUGCGA--GCGACUCCACUGCUCUGCUCUGCCCUG .....(((.((((..((((((((((.....(((....))).(((((.......)))))((((.......)))).......))))).)))))--)))).)))...((........)).... ( -30.40) >DroEre_CAF1 14264 104 + 1 CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUGCGACUGCGACUCCUCU---------------- .....(((.((((((((((((((((.....(((....))).(((((.......)))))((((.......)))).......))))).))))))))))).)))...---------------- ( -39.00) >DroYak_CAF1 14971 112 + 1 CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUGCGA--GCGACUCCUCA-CUCUCCUCC-----G .....(((.((((..((((((((((.....(((....))).(((((.......)))))((((.......)))).......))))).)))))--)))).)))...-.........-----. ( -29.30) >consensus CAAACGGAAUCGCAGUUGCAUCGGCUCUGUUUGUGAACAACUCGAAUUACCAUUUCGAUUCAGUUUUAAUGAGAUUUGUCGCCGACUGCGA__GCGACUCCACU_CUCU_CUCU_____G .....(((.((((..((((((((((..((((....))))..(((((.......)))))((((.......)))).......))))).)))))..)))).)))................... (-22.78 = -24.78 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:45 2006