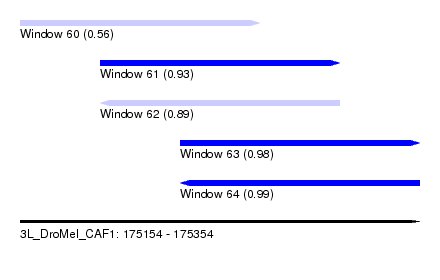
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 175,154 – 175,354 |
| Length | 200 |
| Max. P | 0.991380 |

| Location | 175,154 – 175,274 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -35.35 |
| Energy contribution | -35.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 175154 120 + 23771897 GAUGAGCUGGCGUCGUCGGUUGAUGCUGAGUCGUAUUAGUAUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGC ....(((((((...)))))))((((((((......))))))))...((((...(((((.......)))))))))((((((....))))))......((((((((((....)))))))))) ( -39.90) >DroSec_CAF1 8860 120 + 1 GAUGAGUUGGCGUCGUCGGUUGCUGCUGAGUCGUAUUAGUAUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGC .((((.(..(((.((.....)).)))..).))))............((((...(((((.......)))))))))((((((....))))))......((((((((((....)))))))))) ( -38.30) >DroYak_CAF1 9075 120 + 1 GAUGAGUUGGCGUUGUCGGUUGAUGCUGCAUCGUAUUGGUAUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACAAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGC ..(.((((((((.((.((((....)))))).)))...(((((((.....(((....)))....))))))).....))))).)..............((((((((((....)))))))))) ( -36.50) >consensus GAUGAGUUGGCGUCGUCGGUUGAUGCUGAGUCGUAUUAGUAUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGC .((((..((((((((.....))))))))..)))).........(((((.(((....)))...)))))(((((((.(.....).)))))))......((((((((((....)))))))))) (-35.35 = -35.47 + 0.11)

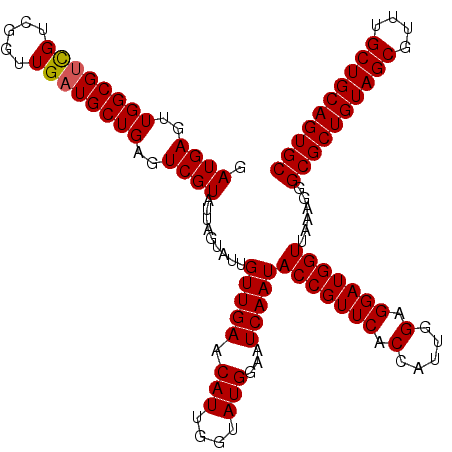
| Location | 175,194 – 175,314 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -39.60 |
| Energy contribution | -39.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 175194 120 + 23771897 AUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUU ..((.(((((...(((((.......)))))))))).))......((((((((...(((((((((((....)))))))))))...((.(((......))).)).))))))))......... ( -40.70) >DroSec_CAF1 8900 120 + 1 AUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUU ..((.(((((...(((((.......)))))))))).))......((((((((...(((((((((((....)))))))))))...((.(((......))).)).))))))))......... ( -40.70) >DroYak_CAF1 9115 120 + 1 AUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACAAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUU (((((.((((...(((((.......)))))))))))))).....((((((((...(((((((((((....)))))))))))...((.(((......))).)).))))))))......... ( -41.50) >consensus AUUGUUGAACAUUGGUAUGGAAUCAAUACCGUUCACCAUUGGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUU .....(((((...(((((.......)))))))))).........((((((((...(((((((((((....)))))))))))...((.(((......))).)).))))))))......... (-39.60 = -39.60 + 0.00)

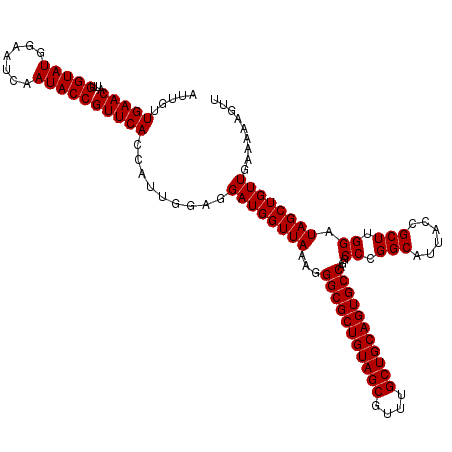
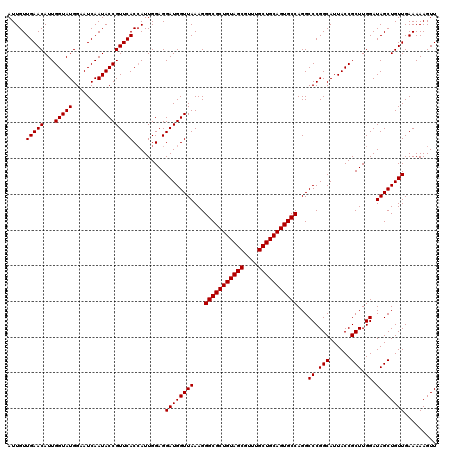
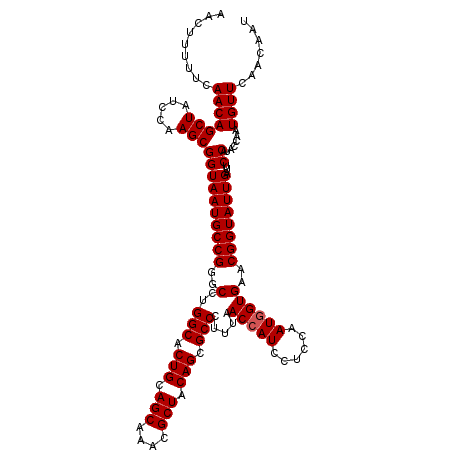
| Location | 175,194 – 175,314 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -29.57 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 175194 120 - 23771897 AACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCCAAUGGUGAACGGUAUUGAUUCCAUACCAAUGUUCAACAAU .........(((((((.....)))((((((((((..(..(((.(((.(((....))).))).))).....(((((......))))))..))))))))...)).......))))....... ( -31.70) >DroSec_CAF1 8900 120 - 1 AACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCCAAUGGUGAACGGUAUUGAUUCCAUACCAAUGUUCAACAAU .........(((((((.....)))((((((((((..(..(((.(((.(((....))).))).))).....(((((......))))))..))))))))...)).......))))....... ( -31.70) >DroYak_CAF1 9115 120 - 1 AACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCCAAUUGUGAACGGUAUUGAUUCCAUACCAAUGUUCAACAAU .............(((.....)))((.....))((....(((.(((.(((....))).))).)))......))........((((((((((((((.......)))))...)))).))))) ( -27.50) >consensus AACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCCAAUGGUGAACGGUAUUGAUUCCAUACCAAUGUUCAACAAU .........(((((((.....)))((((((((((..(..(((.(((.(((....))).))).))).....(((((......))))))..))))))))...)).......))))....... (-29.57 = -29.90 + 0.33)

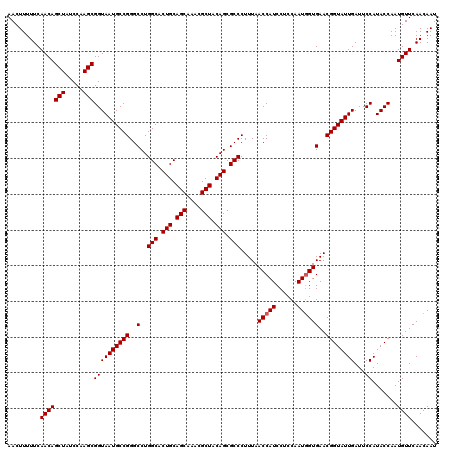
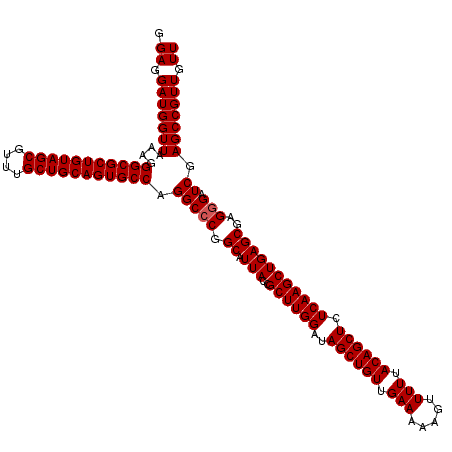
| Location | 175,234 – 175,354 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -51.27 |
| Consensus MFE | -48.57 |
| Energy contribution | -48.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 175234 120 + 23771897 GGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUUUUUACAGCUCUCAAGCUGAGCGAGGGAUCGAGCCGUUGUU .((.(((((((....(((((((((((....))))))))))).(((((.((.(((..((((((..((((((.(((.....))).)))))).)))))))))))..))).)).))))))).)) ( -51.10) >DroSec_CAF1 8940 120 + 1 GGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUUUUUACAGCUCUCAAGCUGAGCGAGGGAUCGAGCCGUUGUU .((.(((((((....(((((((((((....))))))))))).(((((.((.(((..((((((..((((((.(((.....))).)))))).)))))))))))..))).)).))))))).)) ( -51.10) >DroYak_CAF1 9155 120 + 1 GGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUUUUUACAGCUCUCAAGCUGAGCGAGAGUUCGAGCCGUUGUU .......((((....(((((((((((....))))))))))).))))((((......((((((..((((((.(((.....))).)))))).))))))(((((....))))).))))..... ( -51.60) >consensus GGAGGAUGGUUAAAGGGCGCUGUAGCGUUUGCUGCAGUGCCAGGCCCGGCAUUACCGCUUGGAUAGCUGUUGAAAAAGUUUUUACAGCUCUCAAGCUGAGCGAGGGAUCGAGCCGUUGUU .((.(((((((....(((((((((((....))))))))))).(((((.((.(((..((((((..((((((.(((.....))).)))))).)))))))))))..))).)).))))))).)) (-48.57 = -48.90 + 0.33)

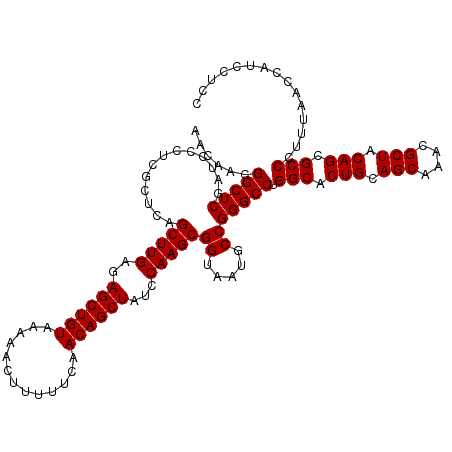
| Location | 175,234 – 175,354 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -37.62 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 175234 120 - 23771897 AACAACGGCUCGAUCCCUCGCUCAGCUUGAGAGCUGUAAAAACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCC ......(((((.............(((((..((((((.............))))))...)))))((.....))))))).(((.(((.(((....))).))).)))............... ( -37.62) >DroSec_CAF1 8940 120 - 1 AACAACGGCUCGAUCCCUCGCUCAGCUUGAGAGCUGUAAAAACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCC ......(((((.............(((((..((((((.............))))))...)))))((.....))))))).(((.(((.(((....))).))).)))............... ( -37.62) >DroYak_CAF1 9155 120 - 1 AACAACGGCUCGAACUCUCGCUCAGCUUGAGAGCUGUAAAAACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCC ......(((((.............(((((..((((((.............))))))...)))))((.....))))))).(((.(((.(((....))).))).)))............... ( -37.62) >consensus AACAACGGCUCGAUCCCUCGCUCAGCUUGAGAGCUGUAAAAACUUUUUCAACAGCUAUCCAAGCGGUAAUGCCGGGCCUGGCACUGCAGCAAACGCUACAGCGCCCUUUAACCAUCCUCC ......(((((.............(((((..((((((.............))))))...)))))((.....))))))).(((.(((.(((....))).))).)))............... (-37.62 = -37.62 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:54 2006