
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,237,199 – 13,237,329 |
| Length | 130 |
| Max. P | 0.995611 |

| Location | 13,237,199 – 13,237,295 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13237199 96 + 23771897 UGAAAUUGAAAUUUAAUUUGCGAAGCGAGCUGGCAAAAAAAAA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUU ..(((((((.................((((((((.........------.....))))))))..........(((......)))...........))))))) ( -22.54) >DroSec_CAF1 30499 84 + 1 UGAAAUUUAAUUUUA-------AAGCGAGCUGGCA-A----AA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUU ((((...........-------....((((((((.-.----..------.....))))))))..........(((......))).....))))......... ( -20.50) >DroSim_CAF1 38106 92 + 1 UGAAAUUGAAAUUUAAUUUGCGAAGCGAGCUGGCGAA----AA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUU ..(((((((.................(((((((((..----..------....)))))))))..........(((......)))...........))))))) ( -26.00) >DroEre_CAF1 20005 91 + 1 UGAAAUUGAAAUUUAAUUUGCGAAGCGAGCUGGCGAAA-----------AAAGCGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCACUUU .....(((((........(((.....(((((((((...-----------....))))))))).......)))(((......))).....)))))........ ( -25.10) >DroYak_CAF1 30687 93 + 1 UGAAAUUGAAAUUUAAUUUGCGAAGCGAGCUGGCGAAA---AA------AAAACGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCACUUU .....(((((........(((.....(((((((((...---..------....))))))))).......)))(((......))).....)))))........ ( -24.40) >DroAna_CAF1 116541 94 + 1 GGA------AAUUUAAUUUGC--AGGGACUUGGCUAACAAAAAAAAACGAAAACGCCAGGUCAUCAUCAGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUU .((------(........(((--...((((((((....................)))))))).......)))(((......))).....))).......... ( -16.05) >consensus UGAAAUUGAAAUUUAAUUUGCGAAGCGAGCUGGCGAAA___AA______AAAGCGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUU ((((......................((((((((....................))))))))..........(((......))).....))))......... (-18.65 = -18.57 + -0.08)


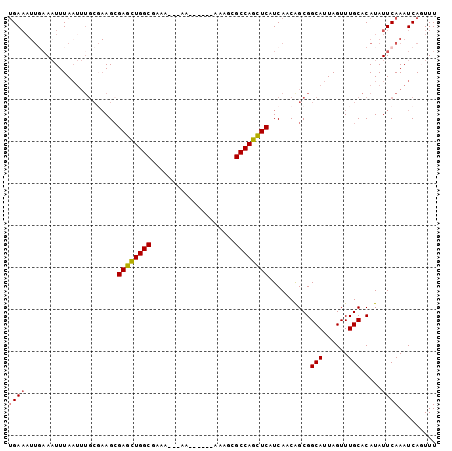
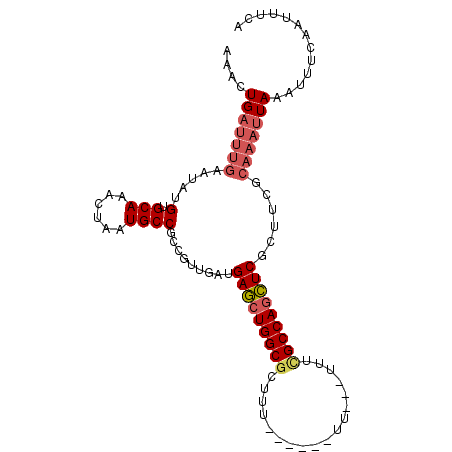
| Location | 13,237,199 – 13,237,295 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -18.40 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13237199 96 - 23771897 AAACUGAUUUGAAUAUGUGCAAACUAAUGCCGCCGUUGAUGAGCUGGCGCUUU------UUUUUUUUUGCCAGCUCGCUUCGCAAAUUAAAUUUCAAUUUCA ....(((((((.....(.(((......))))..((..(.((((((((((....------........)))))))))))..)))))))))............. ( -22.30) >DroSec_CAF1 30499 84 - 1 AAACUGAUUUGAAUAUGUGCAAACUAAUGCCGCCGUUGAUGAGCUGGCGCUUU------UU----U-UGCCAGCUCGCUU-------UAAAAUUAAAUUUCA .....(((((((......((....(((((....)))))..(((((((((....------..----.-)))))))))))..-------.....)))))))... ( -20.22) >DroSim_CAF1 38106 92 - 1 AAACUGAUUUGAAUAUGUGCAAACUAAUGCCGCCGUUGAUGAGCUGGCGCUUU------UU----UUCGCCAGCUCGCUUCGCAAAUUAAAUUUCAAUUUCA ....(((((((.....(.(((......))))..((..(.((((((((((....------..----..)))))))))))..)))))))))............. ( -24.70) >DroEre_CAF1 20005 91 - 1 AAAGUGAUUUGAAUAUGUGCAAACUAAUGCCGCUGUUGAUGAGCUGGCGCUUU-----------UUUCGCCAGCUCGCUUCGCAAAUUAAAUUUCAAUUUCA ....(((((((((((((.(((......))))).))))...(((((((((....-----------...)))))))))......)))))))............. ( -24.80) >DroYak_CAF1 30687 93 - 1 AAAGUGAUUUGAAUAUGUGCAAACUAAUGCCGCUGUUGAUGAGCUGGCGUUUU------UU---UUUCGCCAGCUCGCUUCGCAAAUUAAAUUUCAAUUUCA ....(((((((((((((.(((......))))).))))...(((((((((....------..---...)))))))))......)))))))............. ( -25.30) >DroAna_CAF1 116541 94 - 1 AAACUGAUUUGAAUAUGUGCAAACUAAUGCCGCUGAUGAUGACCUGGCGUUUUCGUUUUUUUUUGUUAGCCAAGUCCCU--GCAAAUUAAAUU------UCC .....((((((.....(.(((......))))(((((..(......((((....)))).....)..)))))))))))...--............------... ( -16.20) >consensus AAACUGAUUUGAAUAUGUGCAAACUAAUGCCGCCGUUGAUGAGCUGGCGCUUU______UU___UUUCGCCAGCUCGCUUCGCAAAUUAAAUUUCAAUUUCA ....(((((((.....(.(((......)))).........(((((((((..................)))))))))......)))))))............. (-18.40 = -19.12 + 0.72)


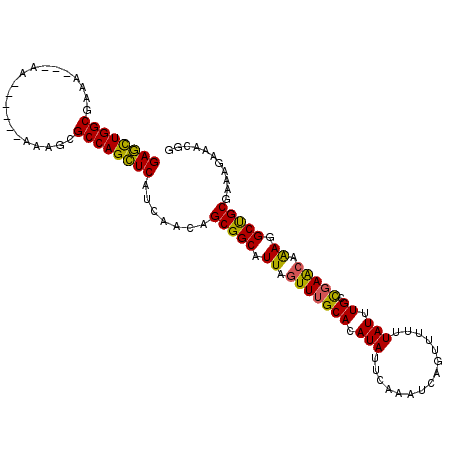
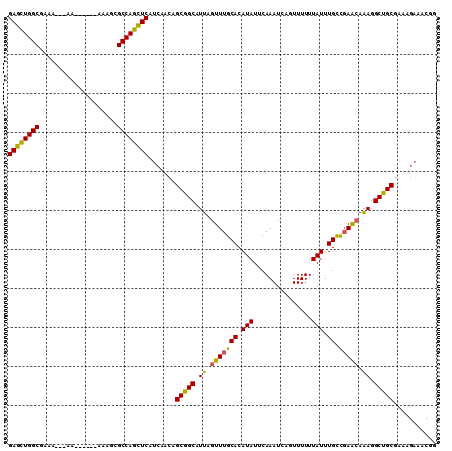
| Location | 13,237,225 – 13,237,329 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -27.56 |
| Energy contribution | -26.93 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13237225 104 + 23771897 GAGCUGGCAAAAAAAAA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUUUUUAUUUGCCGAACAAAGGCUGCUAAAGAAACGG ((((((((.........------.....))))))))......((((((....(((((((.(((...............))).)).)))))....)))))).......... ( -29.20) >DroSec_CAF1 30518 99 + 1 GAGCUGGCA-A----AA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUUUUUAUUUGCCGAACAAAGGCUGCGAAAGAAACGG ((((((((.-.----..------.....)))))))).......(((((....(((((((.(((...............))).)).)))))....)))))........... ( -28.76) >DroSim_CAF1 38132 100 + 1 GAGCUGGCGAA----AA------AAAGCGCCAGCUCAUCAACGGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUUUUUAUUUGCCGAACAAAGGCUGCGAAAGAAACGG (((((((((..----..------....))))))))).......(((((....(((((((.(((...............))).)).)))))....)))))........... ( -31.76) >DroEre_CAF1 20031 99 + 1 GAGCUGGCGAAA-----------AAAGCGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCACUUUUUUAUUUGCCGAACAAAGGCUGCGAAAGAAACGG (((((((((...-----------....))))))))).......(((((....(((((((.(((...............))).)).)))))....)))))........... ( -31.66) >DroYak_CAF1 30713 101 + 1 GAGCUGGCGAAA---AA------AAAACGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCACUUUUUUAUUUGCCGAACAAAGGCUGCGAAAGAAACGG (((((((((...---..------....))))))))).......(((((....(((((((.(((...............))).)).)))))....)))))........... ( -30.96) >DroAna_CAF1 116559 100 + 1 GACUUGGCUAACAAAAAAAAACGAAAACGCCAGGUCAUCAUCAGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUUUUUAUUUGUUCAGAAGAGGCCGCG---------- ((((((((....................)))))))).......(((((.....((((.(((.((..(((.....)))..)).))).))))....))))).---------- ( -27.15) >consensus GAGCUGGCGAAA___AA______AAAGCGCCAGCUCAUCAACAGCGGCAUUAGUUUGCACAUAUUCAAAUCAGUUUUUUAUUUGCCGAACAAAGGCUGCGAAAGAAACGG ((((((((....................)))))))).......(((((.((.(((((((.(((...............))).)).))))).)).)))))........... (-27.56 = -26.93 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:00 2006