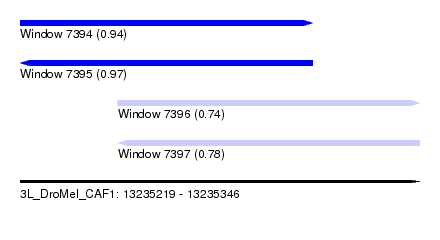
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,235,219 – 13,235,346 |
| Length | 127 |
| Max. P | 0.965221 |

| Location | 13,235,219 – 13,235,312 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13235219 93 + 23771897 AGCCAAAAACUAGUUUAUGGUCAACGGUUUUUUU------UUUUUUCGGAUGCAUAAUUCGCUCAAAAUAGCUGACCAAAUGACUUUUAGUUCGGCUCA ((((...((((((.....(((((..((((..((.------((((..(((((.....)))))...)))).))..))))...))))).)))))).)))).. ( -20.00) >DroSec_CAF1 28596 88 + 1 AGCCAAAAACUAGUUUAUGGUCAACGCAUU----------UUUUUUCGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUU-UUAUUCGGCUCA ((((.......((((..((((((..(((((----------(......)))))).......(((......)))))))))...)))).-......)))).. ( -22.74) >DroSim_CAF1 33561 89 + 1 AGCCAAAAACUAGUUUAUGGUCAACGCAUU----------UUUUUUCGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUUAUUCGGCUCA ((((((.((..((((..((((((..(((((----------(......)))))).......(((......)))))))))...))))..)).)).)))).. ( -22.40) >DroEre_CAF1 18075 87 + 1 AGCCAAAAACUAGUUUAUGGUCAACGCUUUUUUUC------------GGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUUGUUCAGCUCA ((((((((...((((..((((((..(((...((((------------((..((.......)))))))).)))))))))...)))))))))....))).. ( -20.10) >DroYak_CAF1 28700 99 + 1 AGCCAAAAACUAGUUUAUGGUCAACGCUUUUUUUCUUCUUUUCUUUGGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUUGUUUAGCUCA (((.(((....((((..((((((..(((...((((.((((......)))).((.......))..)))).)))))))))...)))).....))).))).. ( -19.50) >consensus AGCCAAAAACUAGUUUAUGGUCAACGCUUUUUUU______UUUUUUCGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUUGUUCGGCUCA ((((.((....((((..((((((........................((((.....))))(((......)))))))))...)))).....)).)))).. (-16.98 = -17.38 + 0.40)

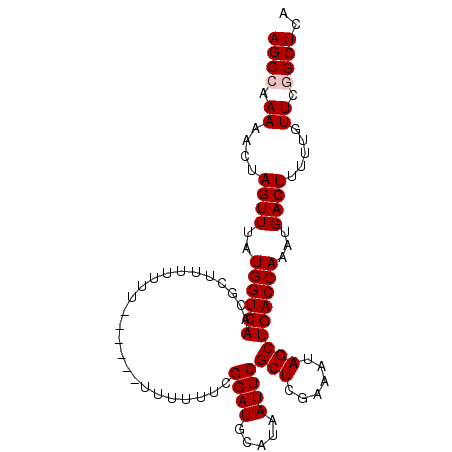
| Location | 13,235,219 – 13,235,312 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13235219 93 - 23771897 UGAGCCGAACUAAAAGUCAUUUGGUCAGCUAUUUUGAGCGAAUUAUGCAUCCGAAAAAA------AAAAAAACCGUUGACCAUAAACUAGUUUUUGGCU ..(((((((..(..(((....((((((((..(((((((((.....))).))........------...))))..))))))))...)))..).))))))) ( -19.20) >DroSec_CAF1 28596 88 - 1 UGAGCCGAAUAA-AAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCGAAAAAA----------AAUGCGUUGACCAUAAACUAGUUUUUGGCU ..(((((((.(.-.(((....((((((((...(((....)))....((((.........----------.))))))))))))...)))..).))))))) ( -24.30) >DroSim_CAF1 33561 89 - 1 UGAGCCGAAUAAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCGAAAAAA----------AAUGCGUUGACCAUAAACUAGUUUUUGGCU ..(((((((.((..(((....((((((((...(((....)))....((((.........----------.))))))))))))...)))..))))))))) ( -23.80) >DroEre_CAF1 18075 87 - 1 UGAGCUGAACAAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCC------------GAAAAAAAGCGUUGACCAUAAACUAGUUUUUGGCU ..(((..((.((..(((....((((((((..(((((.(((.....)))...)------------))))......))))))))...)))..))))..))) ( -20.30) >DroYak_CAF1 28700 99 - 1 UGAGCUAAACAAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCCAAAGAAAAGAAGAAAAAAAGCGUUGACCAUAAACUAGUUUUUGGCU ..(((((((.((..(((....((((((((........(((.....))).((.....))................))))))))...)))..))))))))) ( -19.10) >consensus UGAGCCGAACAAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCGAAAAAA______AAAAAAAGCGUUGACCAUAAACUAGUUUUUGGCU ..(((((((.....(((....((((((((...(((....)))....((........................))))))))))...)))....))))))) (-18.58 = -18.22 + -0.36)


| Location | 13,235,250 – 13,235,346 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -21.37 |
| Consensus MFE | -12.33 |
| Energy contribution | -12.69 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13235250 96 + 23771897 UUU--------UUUUUU-------CGGAUGCAUAAUUCGCUCAAAAUAGCUGACCAAAUGACUUUUA---GUUCGGCUCACUCAUUUUGAGCUCAUAACAAAUGAGAGCAAAGU (((--------(.((((-------(((((.....))))((((((((((((((((.(((.....))).---).)))))).....)))))))))...........))))).)))). ( -20.80) >DroSec_CAF1 28626 92 + 1 -----------UUUUUU-------CGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUU-UU---AUUCGGCUCACUCAUUUCGAGCUCAUAACAAAUGAGAGCAAAGU -----------......-------....(((.......(((((((((((.(((((.(((((...-))---))).)).))))).)))))))))((((.....))))..))).... ( -22.60) >DroSim_CAF1 33591 93 + 1 -----------UUUUUU-------CGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUU---AUUCGGCUCACUCAUUUCGAGCUCAUAACAAAUGAGAGCAAGAU -----------......-------....(((.......(((((((((((.(((((.(((((....))---))).)).))))).)))))))))((((.....))))..))).... ( -23.40) >DroEre_CAF1 18106 90 + 1 UUUC---------------------GGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUU---GUUCAGCUCACUCAUUUUGUGCUCAUAACAAAUGAGAGCACAGU ((((---------------------((..((.......)))))))).((((((.((((......)))---).))))))........((((((((...........)))))))). ( -24.40) >DroYak_CAF1 28731 102 + 1 UUUCUU--CUUUUCUUU-------GGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUU---GUUUAGCUCACUCAUUUUGUGCUCAUAACAAAUGAGAGCAAAGU ......--.....((((-------.((((.....))))((((.....((((((.((((......)))---).))))))...((((((.((.......)))))))))))))))). ( -21.10) >DroAna_CAF1 114825 107 + 1 UUUCAGCUUUUUUCUUUAUACACACAUAUGCAUAAUUCUCCCGAACUAGCUGACCAAAUGACUUUUUCGAGUUCGGCUUAA-------AUGCUCAUAAUAAAUGAGAGCAAAGU .....(((((.(((((.............((((.......(((((((......................))))))).....-------))))...........))))).))))) ( -15.90) >consensus UUU________UUUUUU_______CGGAUGCAUAAUUCGCUCGAAAUAGCUGACCAAAUGACUUUUU___GUUCGGCUCACUCAUUUUGAGCUCAUAACAAAUGAGAGCAAAGU ............................(((..........((((((((((((...................)))))).....))))))..(((((.....))))).))).... (-12.33 = -12.69 + 0.36)


| Location | 13,235,250 – 13,235,346 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13235250 96 - 23771897 ACUUUGCUCUCAUUUGUUAUGAGCUCAAAAUGAGUGAGCCGAAC---UAAAAGUCAUUUGGUCAGCUAUUUUGAGCGAAUUAUGCAUCCG-------AAAAAA--------AAA ....(((..((((.....))))(((((((((.(((..((((((.---.........))))))..)))))))))))).......)))....-------......--------... ( -23.10) >DroSec_CAF1 28626 92 - 1 ACUUUGCUCUCAUUUGUUAUGAGCUCGAAAUGAGUGAGCCGAAU---AA-AAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCG-------AAAAAA----------- ....(((..((((.....))))(((((((((.(((..(((((((---..-.....)))))))..)))))))))))).......)))....-------......----------- ( -26.80) >DroSim_CAF1 33591 93 - 1 AUCUUGCUCUCAUUUGUUAUGAGCUCGAAAUGAGUGAGCCGAAU---AAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCG-------AAAAAA----------- ....(((..((((.....))))(((((((((.(((..(((((((---........)))))))..)))))))))))).......)))....-------......----------- ( -27.10) >DroEre_CAF1 18106 90 - 1 ACUGUGCUCUCAUUUGUUAUGAGCACAAAAUGAGUGAGCUGAAC---AAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCC---------------------GAAA ..(((((((...........)))))))...((((..((((((.(---(((......)))).))))))..)))).(((.....)))....---------------------.... ( -23.40) >DroYak_CAF1 28731 102 - 1 ACUUUGCUCUCAUUUGUUAUGAGCACAAAAUGAGUGAGCUAAAC---AAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCC-------AAAGAAAAG--AAGAAA ..((((((((((((..((.((....)).))..)))))(((...(---(((......))))...)))......)))))))...........-------.........--...... ( -18.90) >DroAna_CAF1 114825 107 - 1 ACUUUGCUCUCAUUUAUUAUGAGCAU-------UUAAGCCGAACUCGAAAAAGUCAUUUGGUCAGCUAGUUCGGGAGAAUUAUGCAUAUGUGUGUAUAAAGAAAAAAGCUGAAA ....(((((...........))))).-------.....(((((..(......)...)))))((((((..(((....)))((((((((....)))))))).......)))))).. ( -23.90) >consensus ACUUUGCUCUCAUUUGUUAUGAGCACAAAAUGAGUGAGCCGAAC___AAAAAGUCAUUUGGUCAGCUAUUUCGAGCGAAUUAUGCAUCCG_______AAAAAA________AAA ..(((((((((((.....))))..........(((..((((((.............))))))..))).....)))))))................................... (-12.59 = -12.92 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:57 2006