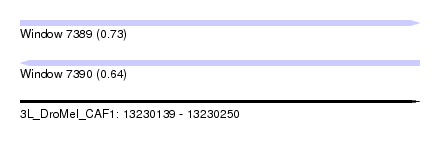
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,230,139 – 13,230,250 |
| Length | 111 |
| Max. P | 0.731476 |

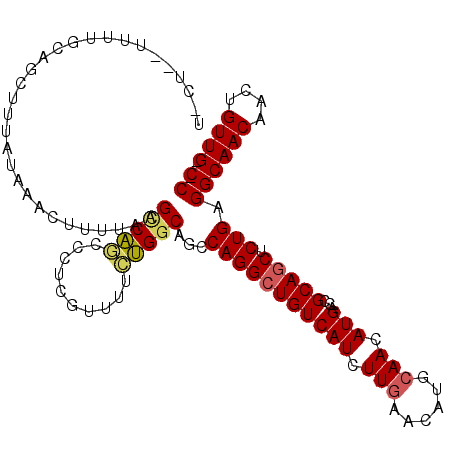
| Location | 13,230,139 – 13,230,250 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13230139 111 + 23771897 UUAU-UUUAUGCUGUUAAAUAAACUUUUAAGCCAGCCCUCGUUUACUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCCGC ....-.....(((.(((((......)))))(((((..........))))))))(((((((((((.(((......))).)))...))))).))).((((((....)))))).. ( -31.70) >DroSec_CAF1 23541 110 + 1 CUCUAGUUUUGCAGCUUUAUAAACUUUAAAGUCAGCCCUCGUUUUCUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCC-- (((.((..((((.((((((.......))))))..((....((((..(((((((...)))))))....))))..)).........))))..)))))(((((....))))).-- ( -28.30) >DroSim_CAF1 28459 110 + 1 CUCUAGUUUUGCAGUUUUAUAAACUUUAAAGUCAGCCCUCGUUUUCUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCC-- (((.((..((((((((.....)))).....((((.....(((.((((((((((...)))))))....))).))).....)))).))))..)))))(((((....))))).-- ( -28.20) >DroEre_CAF1 13048 101 + 1 U---------UCAGCUCCAUAAACUUUUAAGCCAGCCCUCGUUUUCUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAACUCUGAGGCAACAACUGUUGCC-- .---------((((................(((((..........)))))......((.(((((.(((......))).))))).))....))))((((((....))))))-- ( -27.80) >DroYak_CAF1 23513 108 + 1 U-CU-UUCUAGCUGCUCUAUAAACUUUUAAGCCAACCCUCGUUUUCUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCC-- .-..-.((.((((((..............((((....((.(((....)))))...))))(((((.(((......))).))))).))))))..))((((((....))))))-- ( -30.10) >DroAna_CAF1 110207 104 + 1 ------CCAGGUAGCUUUCAGAACUCUGGAGGCGACCCUCUUUCCUCGGCAGUCAGGCUGUCAUCUUCAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCC-- ------...(((.((((((((....)))))))).))).......(((((.(((...((.(((((.(((.....).)).))))).)).))))))))(((((....))))).-- ( -35.90) >consensus U_CU__UUUUGCAGCUUUAUAAACUUUUAAGCCAGCCCUCGUUUUCUGGCAGCCAGGCUGUCAUCUUGAACAUGCAACAUGACCGCAGCUCUGAGGCAACAACUGUUGCC__ ..............................(((((..........)))))...(((((((((((.(((......))).)))...))))).))).((((((....)))))).. (-23.47 = -23.47 + 0.00)

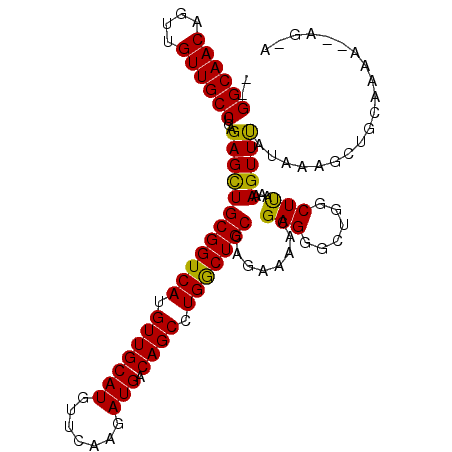
| Location | 13,230,139 – 13,230,250 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -28.42 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13230139 111 - 23771897 GCGGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGUAAACGAGGGCUGGCUUAAAAGUUUAUUUAACAGCAUAAA-AUAA ..((((((....)))))).(((.(((...(((((.(((......))).)))))))))))(((((((((........))))))......(((.....)))))).....-.... ( -35.70) >DroSec_CAF1 23541 110 - 1 --GGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGAAAACGAGGGCUGACUUUAAAGUUUAUAAAGCUGCAAAACUAGAG --((((((....))))))....(((((((((((.(((((((.......))).)))).)))))))....((((.((((....))))...))))....))))............ ( -30.90) >DroSim_CAF1 28459 110 - 1 --GGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGAAAACGAGGGCUGACUUUAAAGUUUAUAAAACUGCAAAACUAGAG --((((((....)))))).(((..(((((((((.(((((((.......))).)))).)))))).))).((((.((((....))))...))))......)))........... ( -29.40) >DroEre_CAF1 13048 101 - 1 --GGCAACAGUUGUUGCCUCAGAGUUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGAAAACGAGGGCUGGCUUAAAAGUUUAUGGAGCUGA---------A --((((((....))))))((((((((((.(((((.(((......))).))))).))..))))(((((..........)))))................))))---------. ( -31.60) >DroYak_CAF1 23513 108 - 1 --GGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGAAAACGAGGGUUGGCUUAAAAGUUUAUAGAGCAGCUAGAA-AG-A --((((((....))))))....(((((((((((.(((((((.......))).)))).)))))(((((..........)))))...............))))))....-..-. ( -35.10) >DroAna_CAF1 110207 104 - 1 --GGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUGAAGAUGACAGCCUGACUGCCGAGGAAAGAGGGUCGCCUCCAGAGUUCUGAAAGCUACCUGG------ --((((((....))))))(((((((((((((((.(((((((.......))).)))).))))))).((((...(.....).))))...))))))))...........------ ( -40.90) >consensus __GGCAACAGUUGUUGCCUCAGAGCUGCGGUCAUGUUGCAUGUUCAAGAUGACAGCCUGGCUGCCAGAAAACGAGGGCUGGCUUAAAAGUUUAUAAAGCUGCAAAA__AG_A ..((((((....))))))...((((((((((((.(((((((.......))).)))).)))))))........(((......)))...))))).................... (-28.42 = -27.87 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:51 2006