| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,224,028 – 13,224,121 |
| Length | 93 |
| Max. P | 0.923127 |

| Location | 13,224,028 – 13,224,121 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
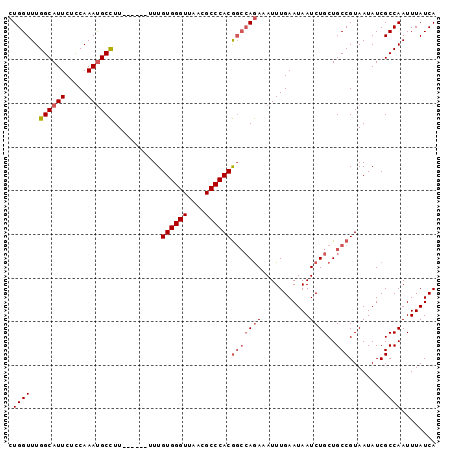
>3L_DroMel_CAF1 13224028 93 + 23771897 CUGGUUUGGCAUUCUCCAAAUGCCUU-----UUUUGUGGGUUAACGCCCACGGCCAGAAAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))..-----....((((((....))))))(((((((...........))))..)))........))))........ ( -25.60) >DroSec_CAF1 17429 92 + 1 CUGGUUUGGCAUUCUCCAAAUGCCUU------UUUGUGGGUUAACGCCCACGGCCAGAAAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))..------...((((((....))))))(((((((...........))))..)))........))))........ ( -25.60) >DroSim_CAF1 22440 90 + 1 CUGGUUUGGCAUUCUCCAAAUGCCUU-------UUGUGGGUUAACGCCCACGGCCAGA-AUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))..-------..((((((....))))))(((((((-..........))))..)))........))))........ ( -25.70) >DroEre_CAF1 7126 92 + 1 CUGGUUUGGCAUUCUUCAAAUGCCUU------UUUGUGGGUUAACGCCCACGGCCAGAUAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))..------...((((((....))))))((((((((.........)))))..)))........))))........ ( -27.10) >DroYak_CAF1 17408 92 + 1 CUGGUUUGGCAUUCUCCAAAUGCCUU------UUUGUGGGUUAACGCCCACGGCCAGAUAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))..------...((((((....))))))((((((((.........)))))..)))........))))........ ( -27.10) >DroAna_CAF1 104913 98 + 1 CUGGUUUGGCAUUCUCCAAACGCUUUUUUUUUUUUGUGGGUUAACGCCCACACGAAAAAAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((.(((((...........(...(((((((.(((((((....))))))).)))))))...)..........))))).......))))........ ( -21.90) >consensus CUGGUUUGGCAUUCUCCAAAUGCCUU______UUUGUGGGUUAACGCCCACGGCCAGAAAUUUGAAUAAUCUGCUGCCGUAAUAUCGCCAAUUUAUCA .((((..((((((.....))))))...........((((((....))))))(((((((...........))))..)))........))))........ (-20.92 = -21.78 + 0.86)


| Location | 13,224,028 – 13,224,121 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
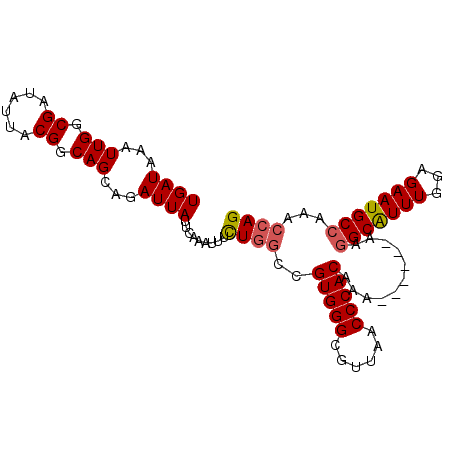
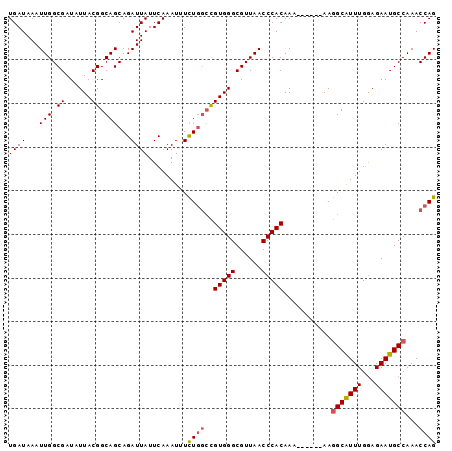
>3L_DroMel_CAF1 13224028 93 - 23771897 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUUUCUGGCCGUGGGCGUUAACCCACAAAA-----AAGGCAUUUGGAGAAUGCCAAACCAG .......((((((...(((((((..((((...........))))))))))).)))))).........-----..(((((((...)))))))....... ( -25.30) >DroSec_CAF1 17429 92 - 1 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUUUCUGGCCGUGGGCGUUAACCCACAAA------AAGGCAUUUGGAGAAUGCCAAACCAG .......((((((...(((((((..((((...........))))))))))).))))))........------..(((((((...)))))))....... ( -25.30) >DroSim_CAF1 22440 90 - 1 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAU-UCUGGCCGUGGGCGUUAACCCACAA-------AAGGCAUUUGGAGAAUGCCAAACCAG .......((((((...(((((((..((((..........-))))))))))).)))))).......-------..(((((((...)))))))....... ( -25.40) >DroEre_CAF1 7126 92 - 1 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUAUCUGGCCGUGGGCGUUAACCCACAAA------AAGGCAUUUGAAGAAUGCCAAACCAG .......((((((...(((((((..(((((.........)))))))))))).))))))........------..(((((((...)))))))....... ( -26.30) >DroYak_CAF1 17408 92 - 1 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUAUCUGGCCGUGGGCGUUAACCCACAAA------AAGGCAUUUGGAGAAUGCCAAACCAG .......((((((...(((((((..(((((.........)))))))))))).))))))........------..(((((((...)))))))....... ( -26.30) >DroAna_CAF1 104913 98 - 1 UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUUUUUUCGUGUGGGCGUUAACCCACAAAAAAAAAAAAGCGUUUGGAGAAUGCCAAACCAG ........((.((......)).))((.((....))...((((((..((((((......))))))..))))))...))((((((......))))))... ( -24.30) >consensus UGAUAAAUUGGCGAUAUUACGGCAGCAGAUUAUUCAAAUUUCUGGCCGUGGGCGUUAACCCACAAA______AAGGCAUUUGGAGAAUGCCAAACCAG ((((...(((.((......)).)))...)))).........((((..(((((......)))))...........(((((((...)))))))...)))) (-21.66 = -21.88 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:44 2006