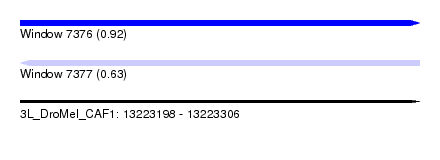
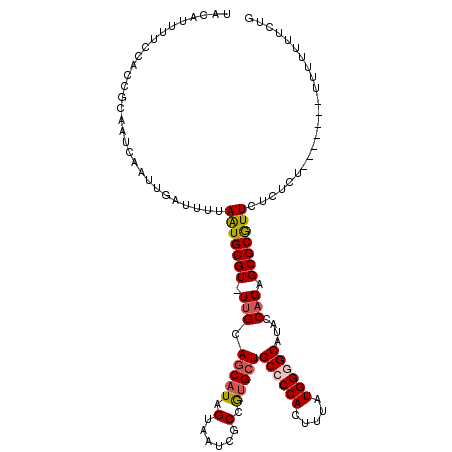
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,223,198 – 13,223,306 |
| Length | 108 |
| Max. P | 0.919642 |

| Location | 13,223,198 – 13,223,306 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13223198 108 + 23771897 CAGAAAAAAA-------AGAGAGAACGCGCUCUGGUAUGCUCCAUAAAGUGGGGCACCAUGGCGAUUACUAUGCUGCAA-ACGCAUUAAAAUCAAUUGAUUGCGGUGGAAAAUGUA ..........-------........((((((.((((..(((((((...))))))))))).)))((((...((((.....-..))))...))))........)))............ ( -28.20) >DroSec_CAF1 16594 108 + 1 CAGAAAAAAA-------AGAGAGAACGCGCUCUGGUAUGCGCCAUAAAGUGGGGCAGCACGGAGAUUACUAUGCUGCGA-ACGCAUUAAAAUCAAUUGAUUGCGGUGGAAAAUGUA ..........-------........((((.(((((((((((((((...)))).((((((..(......)..))))))..-.)))))....))))...)).))))............ ( -25.20) >DroEre_CAF1 6419 108 + 1 CAGAAAAAA--------AGAGAGAAUGCGCUCUGGUAUGCCCCAUAAAGUGGGGCAGCACGGCGAUUACUAUGCUGCAAAACGCACUAAAGUCAAUUGAUUGCGGUGGAAAAUGCA .........--------..((((......)))).(((((((((((...)))))))..(((.(((((((((.((.(((.....))).)).))).....)))))).)))....)))). ( -31.60) >DroYak_CAF1 16517 115 + 1 CAGCAAAAAAAAAAAAUAAAGAGAACGCGCUCUGGUAUGCCCCAUAAAGUGGGGCAGCGCGGCGAUUACUAUGCUGCAA-ACGCAUUAAAAUCAAUUGAUUGCGGUGGAGAAUGCA ..((...............((((......))))....((((((((...))))))))))((((((.......))))))..-..(((((....((.((((....)))).)).))))). ( -33.80) >consensus CAGAAAAAAA_______AGAGAGAACGCGCUCUGGUAUGCCCCAUAAAGUGGGGCAGCACGGCGAUUACUAUGCUGCAA_ACGCAUUAAAAUCAAUUGAUUGCGGUGGAAAAUGCA ...................((((......))))....((((((((...)))))))).(((.(((((((..((((........))))..........))))))).)))......... (-24.45 = -24.70 + 0.25)

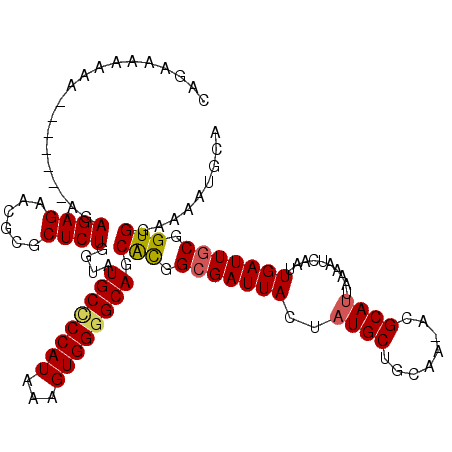
| Location | 13,223,198 – 13,223,306 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13223198 108 - 23771897 UACAUUUUCCACCGCAAUCAAUUGAUUUUAAUGCGU-UUGCAGCAUAGUAAUCGCCAUGGUGCCCCACUUUAUGGAGCAUACCAGAGCGCGUUCUCUCU-------UUUUUUUCUG ............(((........((((...((((..-.....))))...))))((..((((((.(((.....))).))..))))..)))))........-------.......... ( -22.40) >DroSec_CAF1 16594 108 - 1 UACAUUUUCCACCGCAAUCAAUUGAUUUUAAUGCGU-UCGCAGCAUAGUAAUCUCCGUGCUGCCCCACUUUAUGGCGCAUACCAGAGCGCGUUCUCUCU-------UUUUUUUCUG ...............((((....))))..(((((((-((((((((..(......)..)))))).........(((......))))))))))))......-------.......... ( -25.10) >DroEre_CAF1 6419 108 - 1 UGCAUUUUCCACCGCAAUCAAUUGACUUUAGUGCGUUUUGCAGCAUAGUAAUCGCCGUGCUGCCCCACUUUAUGGGGCAUACCAGAGCGCAUUCUCUCU--------UUUUUUCUG (((..........))).............(((((((((((.((((..((....))..))))((((((.....))))))....)))))))))))......--------......... ( -31.10) >DroYak_CAF1 16517 115 - 1 UGCAUUCUCCACCGCAAUCAAUUGAUUUUAAUGCGU-UUGCAGCAUAGUAAUCGCCGCGCUGCCCCACUUUAUGGGGCAUACCAGAGCGCGUUCUCUUUAUUUUUUUUUUUUGCUG .............((((......((((...((((..-.....))))...))))..((((((((((((.....)))))).......))))))...................)))).. ( -29.31) >consensus UACAUUUUCCACCGCAAUCAAUUGAUUUUAAUGCGU_UUGCAGCAUAGUAAUCGCCGUGCUGCCCCACUUUAUGGGGCAUACCAGAGCGCGUUCUCUCU_______UUUUUUUCUG .............................(((((((.(((.(((((.(......).)))))((((((.....))))))....))).)))))))....................... (-18.64 = -19.32 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:39 2006