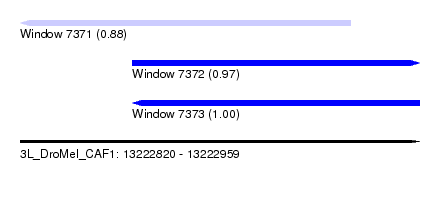
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,222,820 – 13,222,959 |
| Length | 139 |
| Max. P | 0.999975 |

| Location | 13,222,820 – 13,222,935 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -10.81 |
| Energy contribution | -14.87 |
| Covariance contribution | 4.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13222820 115 - 23771897 AAAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUACA-AAAAAGAGGAUCA-AUAAAAACGAGCGUUGCAACACAACAAUUUAUUCU ..............(((...((((.(((.(((((((((((...))))))))))).))).(((((...-....)))))....-..........))))))).................. ( -26.70) >DroSec_CAF1 16218 114 - 1 -AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAGCGCGAUCUAUUUCGCUAUCUCCAUA-AAAAAGAGGAUCA-AUAAAAACGAGCGUUGCAACACAACAAUUUAUUCC -.............(((...((((.(((.(((((((((((...))))))))))).))).((((....-.....))))....-..........))))))).................. ( -24.60) >DroSim_CAF1 21320 114 - 1 -AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAGCGCGAUCUAUUUCGCUAUCUCCAUA-AAAAAGAGGAUCA-AUAAAAACGAGCGUUUCAACACAACAAUUUAUUCU -........((((.((.........(((.(((((((((((...))))))))))).))).((((....-.....))))....-..........)).)))).................. ( -21.60) >DroEre_CAF1 6081 115 - 1 -AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUGCA-AAAAGGAGGAUCGGAAAAAAACGAGCGUUGCAACACAACAAUUUAUAGC -.............(((...((((.(((.(((((((((((...))))))))))).))).(((((...-....))))).(((........)))))))))).................. ( -28.80) >DroWil_CAF1 181629 88 - 1 ----CAUAAGAGAAACAAUUACAAAAAACAAUUCGAUAGAUGCGAGAG------------------U-AAAAGGAAAAUAA-UAA--UAAAAGCGUAAGAAUACAA---UUCCAUUU ----..............((((..........((....))(((....)------------------)-)............-...--.......))))........---........ ( -4.40) >DroYak_CAF1 16153 115 - 1 -AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGUUAUCUCUAUACAAAAAGAGGACCA-AUAAAAACGAGCCGUGCAACACAACAAUUUAUUCA -.............(((.....((.(((.(((((((((((...))))))))))).))).(((((........)))))....-..........))..))).................. ( -20.90) >consensus _AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUAUA_AAAAAGAGGAUCA_AUAAAAACGAGCGUUGCAACACAACAAUUUAUUCU ..............(((...((((.(((.(((((((((((...))))))))))).))).((((..........))))...............))))))).................. (-10.81 = -14.87 + 4.06)

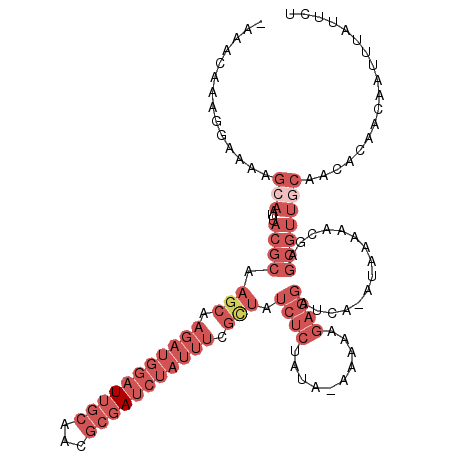

| Location | 13,222,859 – 13,222,959 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13222859 100 + 23771897 CCUCUUUUU-UGUAGAGAUAGCGAAAUAGAUCGCGUUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUUUUUUUUUGUGCAGGCAUUUUUCGCU .((((....-...))))..(((((((..(((.((...)).))).....(((((((((((......................)))))))))))..))))))) ( -21.25) >DroSec_CAF1 16257 97 + 1 CCUCUUUUU-UAUGGAGAUAGCGAAAUAGAUCGCGCUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUU---UUUUGUGCAGGCAUUUUUCGCU .((((....-...))))..(((((((..(((.((...)).))).....(((((((((((..((.........))..---..)))))))))))..))))))) ( -22.60) >DroSim_CAF1 21359 97 + 1 CCUCUUUUU-UAUGGAGAUAGCGAAAUAGAUCGCGCUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUU---UUUUGUGCAGGCAUUUUUCGCU .((((....-...))))..(((((((..(((.((...)).))).....(((((((((((..((.........))..---..)))))))))))..))))))) ( -22.60) >DroEre_CAF1 6121 96 + 1 CCUCCUUUU-UGCAGAGAUAGCGAAAUAGAUCGCGUUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUU----UUUGUGCAUGCUUUUUUCGCU .........-.((..(((.(((((((..(((..(((.((((......))))..)))..)))...))))...(((..----.....))).))).)))..)). ( -19.30) >DroYak_CAF1 16192 98 + 1 CCUCUUUUUGUAUAGAGAUAACGAAAUAGAUCGCGUUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUU---UUUUGUGCAUGCAUUUUUCGCU ........((((((((((.(((((((..(((..(((.((((......))))..)))..)))...))))....))))---))))))))).((.......)). ( -19.30) >consensus CCUCUUUUU_UAUAGAGAUAGCGAAAUAGAUCGCGUUGCAAUCCAUCUUGCUUGCGUAAUUGCUUUUCCUUUGUUU___UUUUGUGCAGGCAUUUUUCGCU .((((........))))..(((((((..(((..(((.((((......))))..)))..)))((......................)).......))))))) (-18.05 = -18.05 + 0.00)

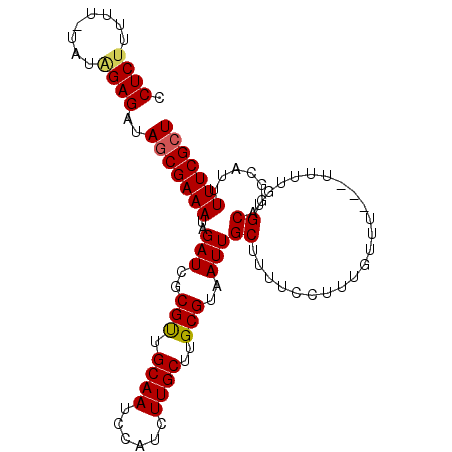
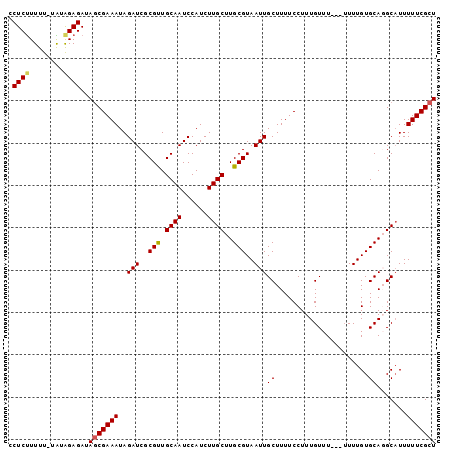
| Location | 13,222,859 – 13,222,959 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.91 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -20.45 |
| Energy contribution | -20.81 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.13 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13222859 100 - 23771897 AGCGAAAAAUGCCUGCACAAAAAAAAAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUACA-AAAAAGAGG (((((((..(((.(((..............................))).)))...((((((((...)))))))))))))))..((((...-....)))). ( -22.61) >DroSec_CAF1 16257 97 - 1 AGCGAAAAAUGCCUGCACAAAA---AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAGCGCGAUCUAUUUCGCUAUCUCCAUA-AAAAAGAGG (((((((..(((.(((......---.....................))).)))...((((((((...)))))))))))))))..(((....-.....))). ( -20.63) >DroSim_CAF1 21359 97 - 1 AGCGAAAAAUGCCUGCACAAAA---AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAGCGCGAUCUAUUUCGCUAUCUCCAUA-AAAAAGAGG (((((((..(((.(((......---.....................))).)))...((((((((...)))))))))))))))..(((....-.....))). ( -20.63) >DroEre_CAF1 6121 96 - 1 AGCGAAAAAAGCAUGCACAAA----AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUGCA-AAAAGGAGG (((((((...((.(((.....----.....................))).))....((((((((...)))))))))))))))..((((...-....)))). ( -22.27) >DroYak_CAF1 16192 98 - 1 AGCGAAAAAUGCAUGCACAAAA---AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGUUAUCUCUAUACAAAAAGAGG (((((((..(((.(((......---.....................))).)))...((((((((...)))))))))))))))..((((........)))). ( -20.63) >consensus AGCGAAAAAUGCCUGCACAAAA___AAACAAAGGAAAAGCAAUUACGCAAGCAAGAUGGAUUGCAACGCGAUCUAUUUCGCUAUCUCUAUA_AAAAAGAGG (((((((..(((.(((..............................))).)))...((((((((...)))))))))))))))..((((........)))). (-20.45 = -20.81 + 0.36)

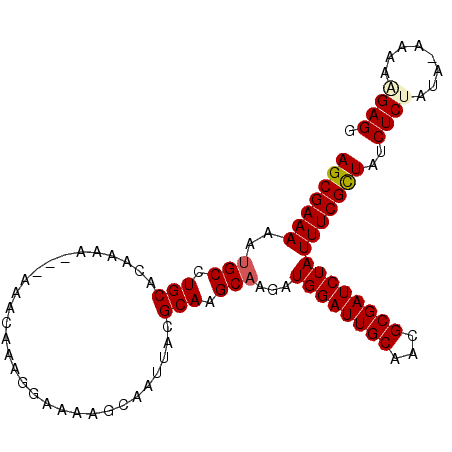
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:35 2006