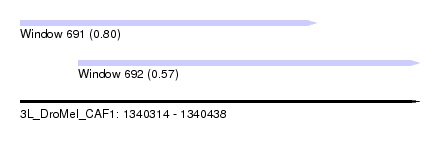
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,340,314 – 1,340,438 |
| Length | 124 |
| Max. P | 0.801439 |

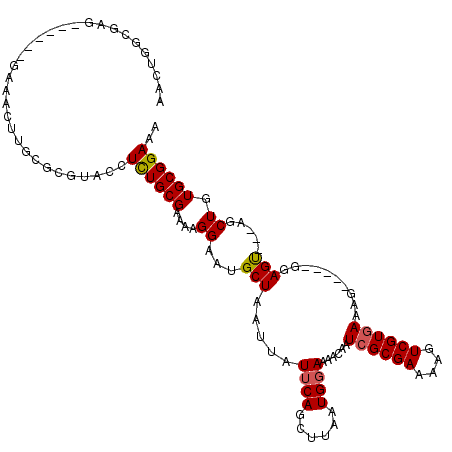
| Location | 1,340,314 – 1,340,406 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1340314 92 + 23771897 AAGCUGAGAAAGCUG----------------------GGAAACUGGCAAG------GAAACUUGCGCGUACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGAUUAAUGGAAAACAAUCGCG .((((.....))))(----------------------.((..((((((((------....)))))((((.(((........))).))))........))).....((.....)).)).). ( -24.40) >DroSec_CAF1 12562 83 + 1 AAG---------CUG----------------------GGAAACUGGCGAG------GAAACUUGCGCGCACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCG (((---------(((----------------------((......(((((------....)))))(((..(((........)))..)))......))))))))................. ( -22.90) >DroYak_CAF1 13551 119 + 1 AAGCUGA-AAAGCUGGGAAAAUGGGGGAAACUGCGGGGGAAACUGGCGAGGAGCAGGAAACUUGCGCGUACCUUUGCGAAGAGGAAUGCUAAUUAUGCAGCUUAAUGGAAAACAAUCGCG .((((..-..)))).......((.((....)).)).((....)).((((.((((((....))...((((.(((((....))))).))))..........))))..((.....)).)))). ( -34.40) >consensus AAGCUGA_AAAGCUG______________________GGAAACUGGCGAG______GAAACUUGCGCGUACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCG .((((.....))))............................((((((((..........)))))((((.(((........))).))))........))).................... (-14.02 = -14.30 + 0.28)

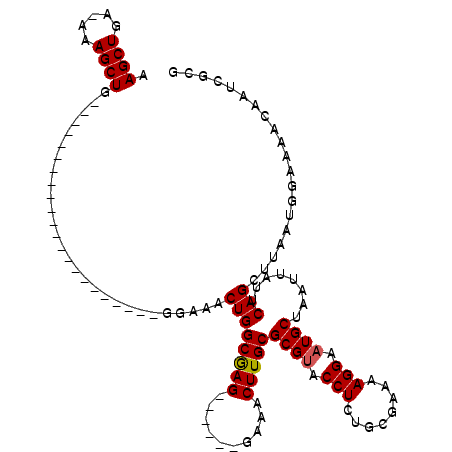
| Location | 1,340,332 – 1,340,438 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.36 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1340332 106 + 23771897 AACUGGCAAG------GAAACUUGCGCGUACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGAUUAAUGGAAAACAAUCGCGAAAAGUCGUAAAAG-----GGAGU---AGCUGUGCGGAAA ..((((((((------....)))))((((.(((........))).))))........)))...............(((((...(((..(.....-----...).---.))).)))))... ( -26.20) >DroSec_CAF1 12571 106 + 1 AACUGGCGAG------GAAACUUGCGCGCACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCGAAAAGUCGUGAAAG-----GGAGU---AGCUGUGCGGAAA .....(((((------....))))).(((((..((((......(((((.....))))).................((((((....))))))...-----...))---))..))))).... ( -31.10) >DroSim_CAF1 12651 95 + 1 AACUGGCGAG------GAAAC-----------UCUGCGAAAAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCGAAGAGUCGUGAAAG-----GGAGU---AGCUGUGCGGAAA ..((((((((------....)-----------)).))..........((((....((((......))))......((((((....))))))...-----....)---)))....)))... ( -21.50) >DroEre_CAF1 12931 100 + 1 --------------------GUUGCACGUACCUUUGCGUAGAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCGAAAAGUCGUGAAAGAGUUGGGAGCUAGGGCUGUGCGGAAA --------------------.(((((((..(((..(((((......)))......((((((((..((.....)).((((((....))))))..)))))))).)).)))..)))))))... ( -28.50) >DroYak_CAF1 13590 112 + 1 AACUGGCGAGGAGCAGGAAACUUGCGCGUACCUUUGCGAAGAGGAAUGCUAAUUAUGCAGCUUAAUGGAAAACAAUCGCGAGAAGUCGUGAAAG-----GGAGU---AGCUGUGCGGAAA ..((((((((..........)))))((((.(((((....))))).)))).......((((((...((.....)).((((((....))))))...-----.....---)))))).)))... ( -30.70) >consensus AACUGGCGAG______GAAACUUGCGCGUACCUCUGCGAAAAGGAAUGCUAAUUAUUCAGCUUAAUGGAAAACAAUCGCGAAAAGUCGUGAAAG_____GGAGU___AGCUGUGCGGAAA ................................((((((....((...(((.....((((......))))......((((((....))))))..........))).....)).)))))).. (-14.36 = -14.36 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:42 2006