| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,221,214 – 13,221,320 |
| Length | 106 |
| Max. P | 0.577506 |

| Location | 13,221,214 – 13,221,320 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13221214 106 + 23771897 --CUCCCGUGGCUCAUUGAUCCACACAGGUCGACGGGUA------AAUUU-CUCGGAUUGGGUUCUCUUCUCUGCGAGCCAGUGGACCCGAUCU--CCA-UCCUCGUCCAACUCG--GUC --...((((((.((...)).))))...((.(((((((..------.....-))))((((((((((.((.(((...)))..)).)))))))))).--...-...))).)).....)--).. ( -33.60) >DroSec_CAF1 14619 106 + 1 --CUCCCCUGGCACAUUUAUCCACACAGGUCGAUGGGUA------AAUUU-CUCGGAUUGGGUUCUCUUCUCCGCGAGCCAGUGGACCCGAUCC--CCA-UUCUCGUCCAACUCG--GUC --......(((.((((((((((((.......).))))))------)))..-...(((((((((((.((.(((...)))..)).)))))))))))--...-.....))))).....--... ( -30.80) >DroSim_CAF1 19724 106 + 1 --CUCCCCUGGCACAUUGAUCCACACAGGUCGAUGGGUA------AAUUU-CUCGGAUUGGGUUCUCUUCUCCGCGAGCCAGUGGACCCGAUCC--CCA-UCCUCGUCCAACUCG--GUC --....((.((..((((((((......))))))))((..------.....-...(((((((((((.((.(((...)))..)).)))))))))))--...-.......))..)).)--).. ( -31.85) >DroEre_CAF1 4439 106 + 1 --CUCCUCCGGCAGAUUGAUCCACACUGGUCGAUGGGUG------AAUUC-CUCGGAGCGGGAAGCCUUCUCCGCGAACCAGUGGAUCCGUUCC--ACA-UCCUCGCCCCACAUU--GUC --.......((((((((((((......)))))))(((((------(....-(.(((((.(((...))).))))).).....(((((.....)))--)).-...))))))....))--))) ( -35.20) >DroYak_CAF1 14513 106 + 1 --CUCAUGCGGCACAUUGACCCAACCUGGUCGAUGGGUG------ACGUU-CUCGGAAUGGGUUUGCUUCUCCGCGAACCAGUGCACCCGCUCC--CCA-UCCUCGACCACCAUU--GGA --..................((((..(((((((.(((((------.((((-(...)))))(((((((......)))))))((((....))))..--.))-))))))))))...))--)). ( -35.20) >DroAna_CAF1 102357 119 + 1 CCCGGCUCUAAGGUGUUGGUCCAAUCGGGCCGACGAGUCUUCCGAGCUUUACUCG-UUUUCGUAUUCCGCUUGGACAUCCAGUGAACCCGCUCGAACGACUCUCCGUCAAACUCAUCGUC ...(((........((((((((....))))))))(((((.(((((((..(((...-.....)))....)))))))..((.((((....)))).))..)))))...)))............ ( -37.70) >consensus __CUCCCCUGGCACAUUGAUCCACACAGGUCGAUGGGUA______AAUUU_CUCGGAUUGGGUUCUCUUCUCCGCGAGCCAGUGGACCCGAUCC__CCA_UCCUCGUCCAACUCG__GUC ..........((.((((((((......)))))))).))................((((((((........(((((......))))))))))))).......................... (-18.39 = -18.95 + 0.56)

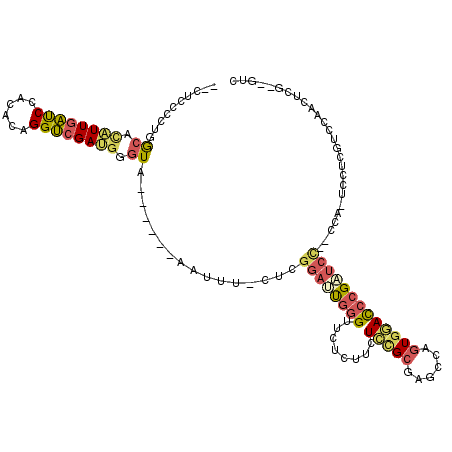
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:33 2006