
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,220,323 – 13,220,423 |
| Length | 100 |
| Max. P | 0.946797 |

| Location | 13,220,323 – 13,220,423 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -14.11 |
| Energy contribution | -15.27 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
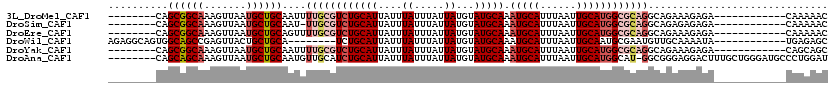
>3L_DroMel_CAF1 13220323 100 + 23771897 GUUUUUG------------UCUCUUUCUGCCUGCGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGACGCAAAAUUGCAGCAUUAACUUUGCCGCUG-------- .......------------.............(((...(((((((...(((.(((((((................))))))).))).)))))))(((.......))))))..-------- ( -20.29) >DroSim_CAF1 18824 99 + 1 GUUUUUG------------UCUCUCUCUGCCUGCGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGACGCAA-AUUGCAGCAUUAACUUUGCCGCUG-------- .......------------.............(((...(((((((...(((.(((((((................))))))).))))-))))))(((.......))))))..-------- ( -20.19) >DroEre_CAF1 3473 100 + 1 GUUUUUG------------UCUCUUUCUGCCUGCGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGACGCAAAACUGCAGCAUUAACUUUGCCGCUG-------- .......------------.............(((....((((...(((((.(((((((................))))))).(((....))).)))))....)))))))..-------- ( -17.99) >DroWil_CAF1 178090 100 + 1 GCUCUCA------------UAUUUUGCAACAUUCGCAUUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGA--------UGCAGCAGUAACUCGGCUGCCACUGCCUCU ((.....------------....((((((........)))))).....(((((((((((................)))))))--------))))(((((......)))))....)).... ( -24.29) >DroYak_CAF1 13565 100 + 1 GCUGCUG------------UCUCUUUCUGCCUGCGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGACGCAAAAUUGCAGCAUUAACUUUGCCGCUG-------- ((.((..------------.........))..))((..(((((((...(((.(((((((................))))))).))).)))))))(((.......))).))..-------- ( -21.19) >DroAna_CAF1 101421 111 + 1 AUCCAGGGCAUCCCAGCAAAGUCCUCCCGCC-AUGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGAUGCAACAUUGCAGCAUUAACUUUGCUGCUG-------- .....(((...)))((((((((.........-((((..((((((....(((((((((((................)))))))))))..))))))))))..))))))))....-------- ( -31.29) >consensus GUUUUUG____________UCUCUUUCUGCCUGCGCCAUGCAAUUAAAUGCAUUUGCAUACAUAAUAAAUAAAUAAUGCAGACGCAAAAUUGCAGCAUUAACUUUGCCGCUG________ ..................................((..((((((....(((.(((((((................))))))).)))..))))))(((.......))).)).......... (-14.11 = -15.27 + 1.17)


| Location | 13,220,323 – 13,220,423 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -19.95 |
| Energy contribution | -21.45 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13220323 100 - 23771897 --------CAGCGGCAAAGUUAAUGCUGCAAUUUUGCGUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCGCAGGCAGAAAGAGA------------CAAAAAC --------..((((((.......))))))...(((((((((((((....((.....))...))))).(((((......)))))))))))))..........------------....... ( -26.60) >DroSim_CAF1 18824 99 - 1 --------CAGCGGCAAAGUUAAUGCUGCAAU-UUGCGUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCGCAGGCAGAGAGAGA------------CAAAAAC --------..((((((.......))))))..(-((((((((((((....((.....))...))))).(((((......)))))))))))))..........------------....... ( -26.70) >DroEre_CAF1 3473 100 - 1 --------CAGCGGCAAAGUUAAUGCUGCAGUUUUGCGUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCGCAGGCAGAAAGAGA------------CAAAAAC --------..((((((.......)))))).(((((...(((((..(((.((.....)).)))(((..(((((......)))))...))).)))))..))))------------)...... ( -28.30) >DroWil_CAF1 178090 100 - 1 AGAGGCAGUGGCAGCCGAGUUACUGCUGCA--------UCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAAUGCGAAUGUUGCAAAAUA------------UGAGAGC ...(((((((((......)))))))))(((--------(.(((((....((.....))...))))).))))......(((((((......)))))))....------------....... ( -28.60) >DroYak_CAF1 13565 100 - 1 --------CAGCGGCAAAGUUAAUGCUGCAAUUUUGCGUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCGCAGGCAGAAAGAGA------------CAGCAGC --------..((((((.......))))))...(((((((((((((....((.....))...))))).(((((......)))))))))))))..........------------....... ( -26.60) >DroAna_CAF1 101421 111 - 1 --------CAGCAGCAAAGUUAAUGCUGCAAUGUUGCAUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCAU-GGCGGGAGGACUUUGCUGGGAUGCCCUGGAU --------...((((((((((.((((((((((..(((((.(((((....((.....))...))))).)))))....)))))..)))))-..(....)))))))))))((....))..... ( -35.20) >consensus ________CAGCGGCAAAGUUAAUGCUGCAAUUUUGCGUCUGCAUUAUUUAUUUAUUAUGUAUGCAAAUGCAUUUAAUUGCAUGGCGCAGGCAGAAAGAGA____________CAAAAAC ..........((((((.......))))))....((((((((((((....((.....))...))))).(((((......)))))))))))).............................. (-19.95 = -21.45 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:32 2006