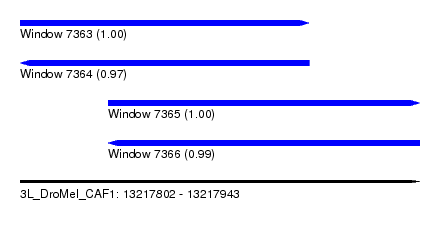
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,217,802 – 13,217,943 |
| Length | 141 |
| Max. P | 0.999535 |

| Location | 13,217,802 – 13,217,904 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13217802 102 + 23771897 GAUAAAUAUAUGAGGAAAAGCCGAGGAAAAACUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUUGUUUGUGCAAACGUAGCAGGAAAUUCAAUA ..............(((...(((((......)))((((((.......))))))......((((.((((((........)))))).)))).))...))).... ( -22.70) >DroSec_CAF1 11169 96 + 1 GAUAAAU--AUGAGGAAAAGCCGAGAAAAAACUCAGAAUGGCAUUUAUGUUCUCCUAGAGCUGAGUUUGCUU----GUGCAAACGAAGCAGGAAAUUCAAUA ....(((--(((((.....((((((......))).....))).)))))))).((((...(((..((((((..----..))))))..)))))))......... ( -22.40) >DroSim_CAF1 16267 96 + 1 GAUAAAU--AUGAGGAAAAGCCGAGGAAAAACUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGAAGCAGGAAAUUCAAUA .......--.....(((...(((((......)))((((((.......))))))......(((..((((((..----..))))))..))).))...))).... ( -22.80) >DroEre_CAF1 1080 96 + 1 GAUAAAU--AUGAAGAAAAGCCGAGGAAAAGCUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGUAGCAGGAAACUCAAUA ....(((--(((((.....((((((......))).....))).))))))))........((((.((((((..----..)))))).))))((....))..... ( -22.10) >DroYak_CAF1 11079 96 + 1 GAUAAAU--AUGAAGAAAAGCCGAGGAAAAACUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGUAGCAGGAAAUUCAAUA .......--.....(((...(((((......)))((((((.......))))))......((((.((((((..----..)))))).)))).))...))).... ( -22.90) >consensus GAUAAAU__AUGAGGAAAAGCCGAGGAAAAACUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU____GUGCAAACGUAGCAGGAAAUUCAAUA ..........((((......(((((......)))((((((.......))))))......(((..((((((........))))))..))).))...))))... (-21.24 = -21.16 + -0.08)

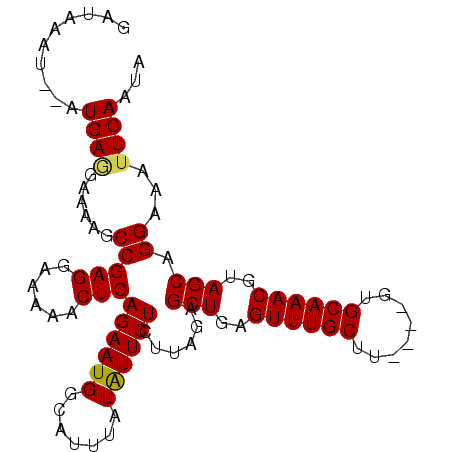
| Location | 13,217,802 – 13,217,904 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.46 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13217802 102 - 23771897 UAUUGAAUUUCCUGCUACGUUUGCACAAACAAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAGUUUUUCCUCGGCUUUUCCUCAUAUAUUUAUC .............(((..((((((........))))))..))).......(((((((.........(((((......)))))..........)))))))... ( -16.71) >DroSec_CAF1 11169 96 - 1 UAUUGAAUUUCCUGCUUCGUUUGCAC----AAGCAAACUCAGCUCUAGGAGAACAUAAAUGCCAUUCUGAGUUUUUUCUCGGCUUUUCCUCAU--AUUUAUC ...(((.(((((((((..((((((..----..))))))..)))...))))))..............(((((......))))).......))).--....... ( -20.60) >DroSim_CAF1 16267 96 - 1 UAUUGAAUUUCCUGCUUCGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAGUUUUUCCUCGGCUUUUCCUCAU--AUUUAUC .............(((..((((((..----..))))))..))).....(((.....(((.(((.....(((......)))))))))..)))..--....... ( -15.60) >DroEre_CAF1 1080 96 - 1 UAUUGAGUUUCCUGCUACGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAGCUUUUCCUCGGCUUUUCUUCAU--AUUUAUC ...((((......(((..((((((..----..))))))..(((((....(((((.........)))))))))).......))).....)))).--....... ( -16.80) >DroYak_CAF1 11079 96 - 1 UAUUGAAUUUCCUGCUACGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAGUUUUUCCUCGGCUUUUCUUCAU--AUUUAUC ...((((((((..(((..((((((..----..))))))..))).....))))..............(((((......)))))......)))).--....... ( -16.10) >consensus UAUUGAAUUUCCUGCUACGUUUGCAC____AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAGUUUUUCCUCGGCUUUUCCUCAU__AUUUAUC ...(((.((((..(((..((((((........))))))..))).....))))..............(((((......))))).......))).......... (-15.46 = -15.46 + -0.00)

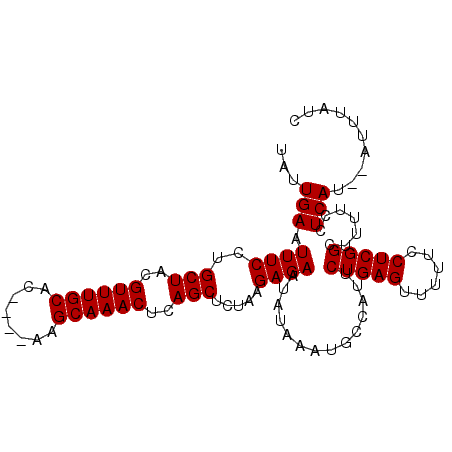
| Location | 13,217,833 – 13,217,943 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13217833 110 + 23771897 CUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUUGUUUGUGCAAACGUAGCAGGAAAUUCAAUAGGAAGAGCGCUGGAAAUUGCCAAAUUAUGAGGAAAUUUA ((((...((((((((.....(((((.....(((((((.(((((((((....))).))))))))))))).....)))))......))).))))).....))))........ ( -28.00) >DroSec_CAF1 11198 106 + 1 CUCAGAAUGGCAUUUAUGUUCUCCUAGAGCUGAGUUUGCUU----GUGCAAACGAAGCAGGAAAUUCAAUAGGAAGAGCGCUGGAAAUUGCCAAAUUAUGAGGAAAUUUA ((((...(((((((((((((((((((..(((..((((((..----..))))))..)))..(.....)..)))).)))))).))))...))))).....))))........ ( -33.20) >DroSim_CAF1 16296 106 + 1 CUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGAAGCAGGAAAUUCAAUAGGAAGAGCGCUGGAAAUUGCCAAAUUAUGAGGAAAUUUA ((((...((((((((.....(((((...(((..((((((..----..))))))..)))..(.....)......)))))......))).))))).....))))........ ( -26.60) >DroEre_CAF1 1109 106 + 1 CUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGUAGCAGGAAACUCAAUAGGAAGGGCGCUGGAAAUUGCCAAAUUAUGAGGAAAUUUA ((((...((((((((.....(((((...((((.((((((..----..)))))).))))((....)).......)))))......))).))))).....))))........ ( -29.20) >DroYak_CAF1 11108 106 + 1 CUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU----GUGCAAACGUAGCAGGAAAUUCAAUAGGAAGGGCGCUGGAAAUUGCCAAAUUAUGAGAAAAUUUA ((((...((((((((.....(((((...((((.((((((..----..)))))).))))..(.....)......)))))......))).))))).....))))........ ( -25.40) >consensus CUCAGAAUGGCAUUUAUAUUCUCUUAGAGCUGAGUUUGCUU____GUGCAAACGUAGCAGGAAAUUCAAUAGGAAGAGCGCUGGAAAUUGCCAAAUUAUGAGGAAAUUUA ((((...((((((((...((((((((..(((..((((((........))))))..)))..(.....)..)))).))))......))).))))).....))))........ (-25.02 = -24.62 + -0.40)



| Location | 13,217,833 – 13,217,943 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13217833 110 - 23771897 UAAAUUUCCUCAUAAUUUGGCAAUUUCCAGCGCUCUUCCUAUUGAAUUUCCUGCUACGUUUGCACAAACAAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAG ........((((.....(((((.((.......(((((.(....)........(((..((((((........))))))..)))...))))).....)).)))))...)))) ( -20.90) >DroSec_CAF1 11198 106 - 1 UAAAUUUCCUCAUAAUUUGGCAAUUUCCAGCGCUCUUCCUAUUGAAUUUCCUGCUUCGUUUGCAC----AAGCAAACUCAGCUCUAGGAGAACAUAAAUGCCAUUCUGAG ........((((.....(((((...........((........)).(((((((((..((((((..----..))))))..)))...)))))).......)))))...)))) ( -24.40) >DroSim_CAF1 16296 106 - 1 UAAAUUUCCUCAUAAUUUGGCAAUUUCCAGCGCUCUUCCUAUUGAAUUUCCUGCUUCGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAG ........((((.....(((((.((.......(((((.(....)........(((..((((((..----..))))))..)))...))))).....)).)))))...)))) ( -20.90) >DroEre_CAF1 1109 106 - 1 UAAAUUUCCUCAUAAUUUGGCAAUUUCCAGCGCCCUUCCUAUUGAGUUUCCUGCUACGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAG ........((((.....(((((............(((......)))((((..(((..((((((..----..))))))..))).....)))).......)))))...)))) ( -19.30) >DroYak_CAF1 11108 106 - 1 UAAAUUUUCUCAUAAUUUGGCAAUUUCCAGCGCCCUUCCUAUUGAAUUUCCUGCUACGUUUGCAC----AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAG ........((((.....(((((.............(((.....)))((((..(((..((((((..----..))))))..))).....)))).......)))))...)))) ( -19.00) >consensus UAAAUUUCCUCAUAAUUUGGCAAUUUCCAGCGCUCUUCCUAUUGAAUUUCCUGCUACGUUUGCAC____AAGCAAACUCAGCUCUAAGAGAAUAUAAAUGCCAUUCUGAG ........((((.....(((((.............(((.....)))((((..(((..((((((........))))))..))).....)))).......)))))...)))) (-19.24 = -19.08 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:29 2006