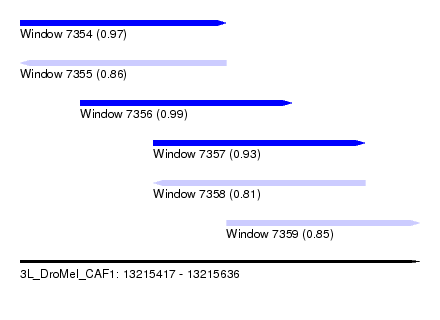
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,215,417 – 13,215,636 |
| Length | 219 |
| Max. P | 0.986816 |

| Location | 13,215,417 – 13,215,530 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -30.34 |
| Energy contribution | -31.02 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215417 113 + 23771897 CUGUGAGCU-------GUGAGUUGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUU ..(((((..-------..((.(((((((.(((........))).)).((((.((((((......)))....))).))))))))).)).)))))....((((((......))))))..... ( -36.30) >DroSec_CAF1 8743 113 + 1 CUGUGAGCU-------GUGAGUUGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUU ..(((((..-------..((.(((((((.(((........))).)).((((.((((((......)))....))).))))))))).)).)))))....((((((......))))))..... ( -36.30) >DroSim_CAF1 13819 113 + 1 CUGUGAGCU-------GUGAGUUGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUU ..(((((..-------..((.(((((((.(((........))).)).((((.((((((......)))....))).))))))))).)).)))))....((((((......))))))..... ( -36.30) >DroYak_CAF1 8681 120 + 1 CUGUGAGCUGCGAGUUGUGAGUUGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUU ..(((....(((((....((.(((((((.(((........))).)).((((.((((((......)))....))).))))))))).)).)))))....((((((......))))))))).. ( -41.40) >DroAna_CAF1 97273 111 + 1 --GCCAGCU-------GUGAGCCGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCGAGCCGGGUCGCGAAGUGUUUCGCCCGUUUUGGCCAUAAAGCCAAACACUU --.......-------((((((.(((((((.((((......((((..((((.((((((......)))....))).))))))))))))))))))).)))(((((......))))).))).. ( -30.60) >consensus CUGUGAGCU_______GUGAGUUGGCGAAAAUAUUAUUUAAUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUU ..(((...........((((((((((((.(((........))).)).((((.((((((......)))....))).)))))))))....)))))....((((((......))))))))).. (-30.34 = -31.02 + 0.68)


| Location | 13,215,417 – 13,215,530 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215417 113 - 23771897 AAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGGCGGCCCGGCUUGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCAACUCAC-------AGCUCACAG .....(((((((....))))))).(((((..(((..(((((((((.(((...((......))...))).)))).((.(((........))).))))))))))..-------.)))))... ( -36.70) >DroSec_CAF1 8743 113 - 1 AAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGGCGGCCCGGCUUGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCAACUCAC-------AGCUCACAG .....(((((((....))))))).(((((..(((..(((((((((.(((...((......))...))).)))).((.(((........))).))))))))))..-------.)))))... ( -36.70) >DroSim_CAF1 13819 113 - 1 AAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGGCGGCCCGGCUUGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCAACUCAC-------AGCUCACAG .....(((((((....))))))).(((((..(((..(((((((((.(((...((......))...))).)))).((.(((........))).))))))))))..-------.)))))... ( -36.70) >DroYak_CAF1 8681 120 - 1 AAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGGCGGCCCGGCUUGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCAACUCACAACUCGCAGCUCACAG ..((((((((((....)))))))....(((((.((.(((((((((.(((...((......))...))).)))).((.(((........))).))))))).))....)))))....))).. ( -39.00) >DroAna_CAF1 97273 111 - 1 AAGUGUUUGGCUUUAUGGCCAAAACGGGCGAAACACUUCGCGACCCGGCUCGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCGGCUCAC-------AGCUGGC-- ..((.(((((((....)))))))))(((((((....)))))...))((((.((.((......))..)).)))).....................((((((....-------.))))))-- ( -33.60) >consensus AAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGGCGGCCCGGCUUGGACUUUAAUUAGCGCCUGGCCUGACAAUUAAAUAAUAUUUUCGCCAACUCAC_______AGCUCACAG ..((((((((((....)))))))...((((((((.......((((.(((...((......))...))).)))).....(((....))).))))))))..................))).. (-29.08 = -29.36 + 0.28)



| Location | 13,215,450 – 13,215,566 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215450 116 + 23771897 AUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAUCUGCAGCAUCUU---CAGU-CCGAUAU .(((...((((.((((((......)))....))).))))..))).....(((..(..((((((......))))))........((((((.(....).))))))..---..).-.)))... ( -30.70) >DroSec_CAF1 8776 113 + 1 AUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GAGCAUCUU---CAGU-CCGAUAU .(((...((((.((((((......)))....))).))))..))).....(((..(..((((((......))))))........(((((((...---)))))))..---..).-.)))... ( -32.30) >DroSim_CAF1 13852 113 + 1 AUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GAGCAUCUU---CAGU-CCGAUAU .(((...((((.((((((......)))....))).))))..))).....(((..(..((((((......))))))........(((((((...---)))))))..---..).-.)))... ( -32.30) >DroYak_CAF1 8721 113 + 1 AUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GACCAUCUU---CAGC-CCGAUAU ..((((.(((..((.(((......)))))..)))((((...................((((((......))))))........((((.((...---)).))))..---..))-)))))). ( -28.10) >DroAna_CAF1 97304 116 + 1 AUUGUCAGGCCAGGCGCUAAUUAAAGUCCGAGCCGGGUCGCGAAGUGUUUCGCCCGUUUUGGCCAUAAAGCCAAACACUUAACGAUGCGGCGA---G-GCAUCUUCUCCAGCACCGAUAU ..((((.(((..((.(((......)))))..))).(((.((((((((((((((((((((((((......))))).........)))).)))))---)-))).))))....))))))))). ( -41.00) >consensus AUUGUCAGGCCAGGCGCUAAUUAAAGUCCAAGCCGGGCCGCCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC___GAGCAUCUU___CAGU_CCGAUAU ..((((.(((((((((((......)))....)))((((.............))))....)))))...................(((((((......)))))))............)))). (-29.40 = -30.04 + 0.64)

| Location | 13,215,490 – 13,215,606 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -27.08 |
| Energy contribution | -28.57 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215490 116 + 23771897 CCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAUCUGCAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCUGGCU ...........(((((.((((((......))))))........((((((.(....).)))))).((((((.((((........)))))))))).))))).(((((....)).))). ( -34.50) >DroSec_CAF1 8816 111 + 1 CCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUCUUGCCGG-- .((((......(((((.((((((......))))))........(((((((...---))))))).((((((.((((........)))))))))).))))).......))))....-- ( -33.72) >DroSim_CAF1 13892 111 + 1 CCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCCGG-- ((.........(((((.((((((......))))))........(((((((...---))))))).((((((.((((........)))))))))).)))))....((....)).))-- ( -33.30) >DroYak_CAF1 8761 107 + 1 CCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC---GACCAUCUUCAGCCCGAUAUUUAUGUUUAUUGACUGGUGAGGCUGCUGCUUCUG------ ...((((((.(......((((((......))))))........((((.((...---)).))))..(((..(((((........))))).)))).))))))..........------ ( -21.70) >consensus CCAAGUCUCUCGCCCAUUUUGGCCAUAAAGCCAAACACUUAACGAUGCUCCAC___GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCCGG__ ..((((..(..(((((.((((((......))))))........(((((((......))))))).((((((.((((........)))))))))).))))).)..))))......... (-27.08 = -28.57 + 1.50)

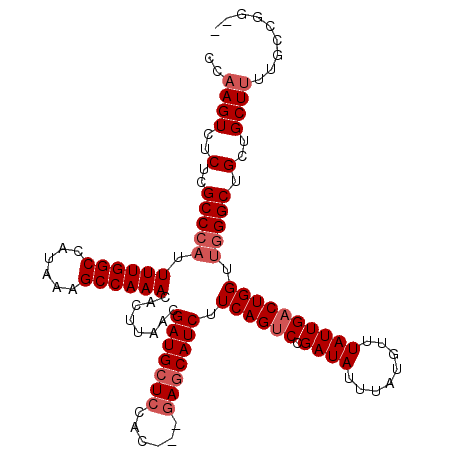
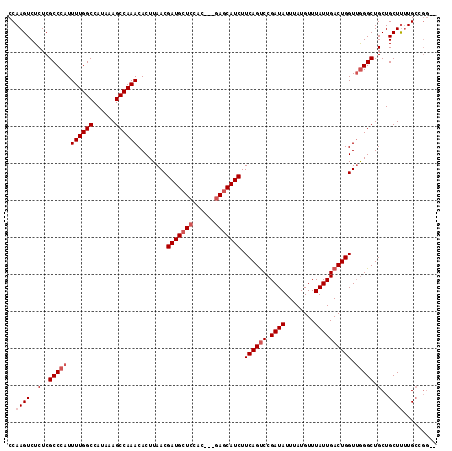
| Location | 13,215,490 – 13,215,606 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -29.61 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215490 116 - 23771897 AGCCAGCAAAAGCAGCAGCCCAACCAGUCAAUAAACAUAAAUAUCGGACUGAAGAUGCUGCAGAUGGAGCAUCGUUAAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGG ..(((((....))....(((((..(((((.(((........)))..)))))..((((((.(....).))))))........(((((((....))))))).)))))........))) ( -35.80) >DroSec_CAF1 8816 111 - 1 --CCGGCAAGAGCAGCAGCCCAACCAGUCAAUAAACAUAAAUAUCGGACUGAAGAUGCUC---GUGGAGCAUCGUUAAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGG --....((((.......(((((..(((((.(((........)))..)))))..(((((((---...)))))))........(((((((....))))))).)))))......)))). ( -35.12) >DroSim_CAF1 13892 111 - 1 --CCGGCAAAAGCAGCAGCCCAACCAGUCAAUAAACAUAAAUAUCGGACUGAAGAUGCUC---GUGGAGCAUCGUUAAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGG --(((((....))....(((((..(((((.(((........)))..)))))..(((((((---...)))))))........(((((((....))))))).)))))........))) ( -34.20) >DroYak_CAF1 8761 107 - 1 ------CAGAAGCAGCAGCCUCACCAGUCAAUAAACAUAAAUAUCGGGCUGAAGAUGGUC---GUGGAGCAUCGUUAAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGG ------..((.((....)).)).((((((......((((((.....((((......))))---((.((((((......)))))).))))))))(((......)))....))).))) ( -26.90) >consensus __CCGGCAAAAGCAGCAGCCCAACCAGUCAAUAAACAUAAAUAUCGGACUGAAGAUGCUC___GUGGAGCAUCGUUAAGUGUUUGGCUUUAUGGCCAAAAUGGGCGAGAGACUUGG .........((((....(((((..(((((.(((........)))..)))))..(((((((......)))))))........(((((((....))))))).)))))....).))).. (-29.61 = -29.80 + 0.19)


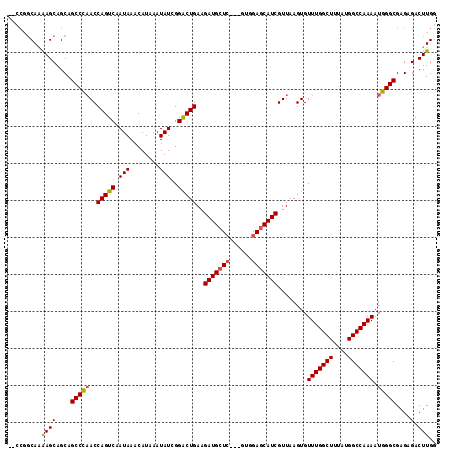
| Location | 13,215,530 – 13,215,636 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -22.92 |
| Energy contribution | -25.05 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13215530 106 + 23771897 AACGAUGCUCCAUCUGCAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCUGGCUGCCAGGCUGGCAACUGACAUCAUCAUCAUC ...((((.((.....(((((((((((((((.((((........))))))))))..))).))))))..((((..(((....)))..))))..))))))......... ( -34.90) >DroSec_CAF1 8856 95 + 1 AACGAUGCUCCAC---GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUCUUGCCGG--------CUGGCAACUGACAUCAUCAUCAUC ...((((.((...---(((((....(((((((((......(((....)))..))))))))).)))))(((((..--------..)))))..))))))......... ( -28.50) >DroSim_CAF1 13932 95 + 1 AACGAUGCUCCAC---GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCCGG--------CUGGCAACUGACAUCAUCAUCAUC ...(((((((...---)))))))..(((((.((((........)))))))))((..(..(((((((......))--------).)))).)..))............ ( -27.70) >DroYak_CAF1 8801 91 + 1 AACGAUGCUCCAC---GACCAUCUUCAGCCCGAUAUUUAUGUUUAUUGACUGGUGAGGCUGCUGCUUCUG------------CUGGCAACUGACAUCAUCAUCAGC ...((((.((...---)).))))........(((..(.(((((..(((.(..(((((((....))))).)------------)..))))..))))).)..)))... ( -21.60) >consensus AACGAUGCUCCAC___GAGCAUCUUCAGUCCGAUAUUUAUGUUUAUUGACUGGUUGGGCUGCUGCUUUUGCCGG________CUGGCAACUGACAUCAUCAUCAUC ...(((((((......)))))))..(((((((((......(((....)))..))))))))).(((..((((((((......))))))))..).))........... (-22.92 = -25.05 + 2.13)

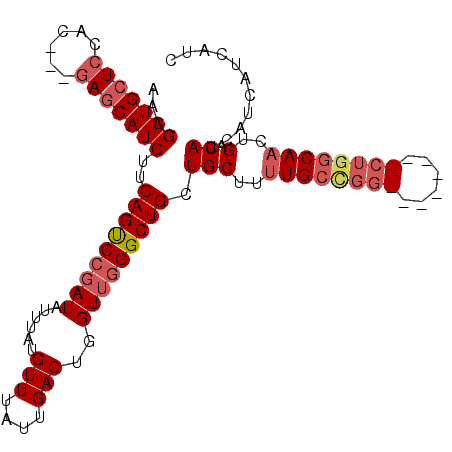
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:22 2006