
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,213,613 – 13,213,754 |
| Length | 141 |
| Max. P | 0.912931 |

| Location | 13,213,613 – 13,213,720 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -35.88 |
| Energy contribution | -35.88 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
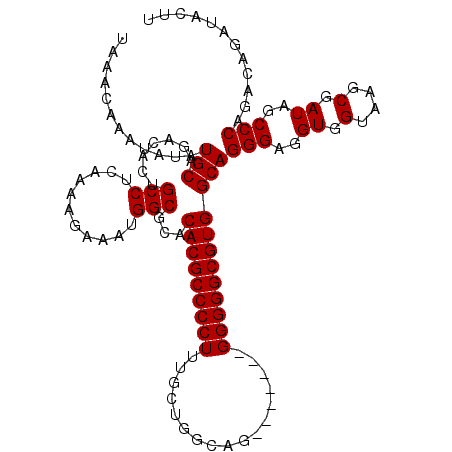
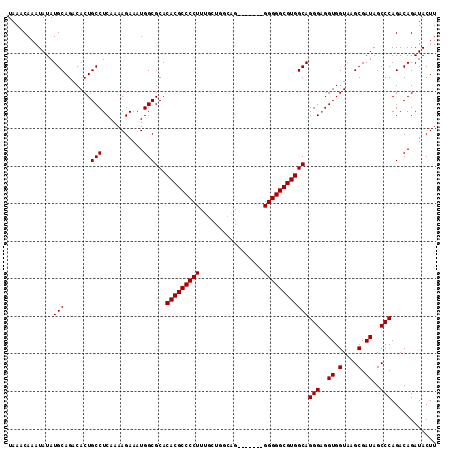
>3L_DroMel_CAF1 13213613 107 - 23771897 AUGGCGCACACGCCCCUUUGCUGGGCGUGGGGCAGGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUUUAUGCCGUACGUGCCAAACAAAACUCC .((((((..(((((((((.(((........))))))))))))((((..(((...((..((....))....)).....)))..))))....))))))........... ( -42.20) >DroSec_CAF1 6954 100 - 1 AUGGCGCACACGCCCCUUUGCUGGCAG-------GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUUUAUGCCGUACGUGCCAAACAAAACUCC .((((((..(((((((((((....)))-------))))))))((((..(((...((..((....))....)).....)))..))))....))))))........... ( -43.10) >DroSim_CAF1 12027 100 - 1 AUGGCGCACACGCCCCUUUGCUGGCUG-------GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUUUAUGCCGUACGUGCCAAACAAAACUCC .((((((..((((((((..(....)..-------))))))))((((..(((...((..((....))....)).....)))..))))....))))))........... ( -39.70) >DroYak_CAF1 6863 98 - 1 AUGGCGCACACGCCCCU-UGCUGUCA--------GGGGGCGUGGCAGGGUGGUGGUAAGCGAUAGCCCAGACAGAUACUUUAUGCCGUACGUGCCAAACAAAACUCC .((((((..((((((((-((....))--------))))))))((((((((.((.(....).)).)))).....((....)).))))....))))))........... ( -40.90) >consensus AUGGCGCACACGCCCCUUUGCUGGCAG_______GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUUUAUGCCGUACGUGCCAAACAAAACUCC .((((((..((((((((.................))))))))(((((((..((.(....).))..))).....((....)).))))....))))))........... (-35.88 = -35.88 + -0.00)

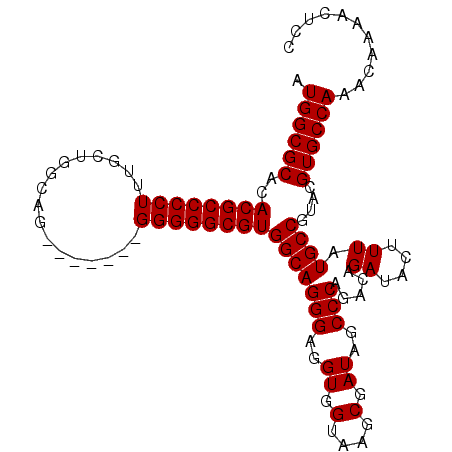
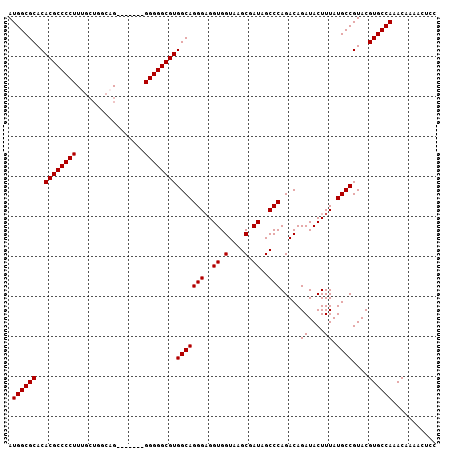
| Location | 13,213,640 – 13,213,754 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -30.31 |
| Energy contribution | -30.31 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13213640 114 + 23771897 AAGUAUCUGUCUGGGCUAUCGCUUACCACCUCCCUGCCACGCCCCCUGCCCCACGCCCAGCAAAGGGGCGUGUGCGCCAUUUCUUUUGAGGCAGUGUCUGCAUAUAUUUGUUUA ((((((.(((.(((((....))))).(((......((((((((((.(((..........)))..)))))))).))(((...((....))))).)))...))).))))))..... ( -33.40) >DroSec_CAF1 6981 107 + 1 AAGUAUCUGUCUGGGCUAUCGCUUACCACCUCCCUGCCACGCCCCC-------CUGCCAGCAAAGGGGCGUGUGCGCCAUUUCUUUUGAGGCAGUGUCUGCAUAUAUUUGUUUA ((((((.(((.(((((....))))).(((......((((((((((.-------.((....))..)))))))).))(((...((....))))).)))...))).))))))..... ( -33.60) >DroSim_CAF1 12054 107 + 1 AAGUAUCUGUCUGGGCUAUCGCUUACCACCUCCCUGCCACGCCCCC-------CAGCCAGCAAAGGGGCGUGUGCGCCAUUUCUUUUGAGGCAGUGUCUGCAUAUAUUUGUUUA ((((((.(((.(((((....))))).(((......((((((((((.-------..(....)...)))))))).))(((...((....))))).)))...))).))))))..... ( -31.30) >DroYak_CAF1 6890 105 + 1 AAGUAUCUGUCUGGGCUAUCGCUUACCACCACCCUGCCACGCCCCC--------UGACAGCA-AGGGGCGUGUGCGCCAUUUCUUUUGAGGCAGUGUCUGCAUAUAUUUGCUUA (((((..(((....((...((((............((((((((((.--------((....))-.)))))))).))(((...((....)))))))))...))..)))..))))). ( -34.30) >consensus AAGUAUCUGUCUGGGCUAUCGCUUACCACCUCCCUGCCACGCCCCC_______CUGCCAGCAAAGGGGCGUGUGCGCCAUUUCUUUUGAGGCAGUGUCUGCAUAUAUUUGUUUA ((((((.(((.(((((....))))).(((......((((((((((...................)))))))).))(((...((....))))).)))...))).))))))..... (-30.31 = -30.31 + 0.00)

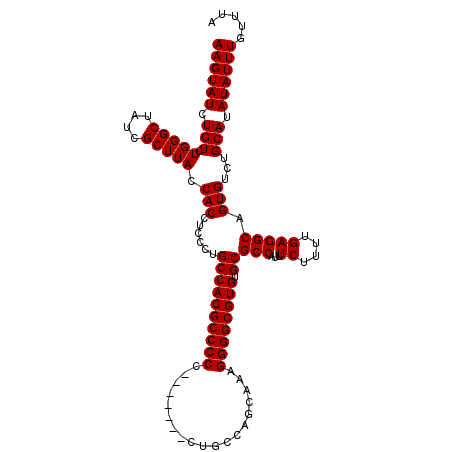
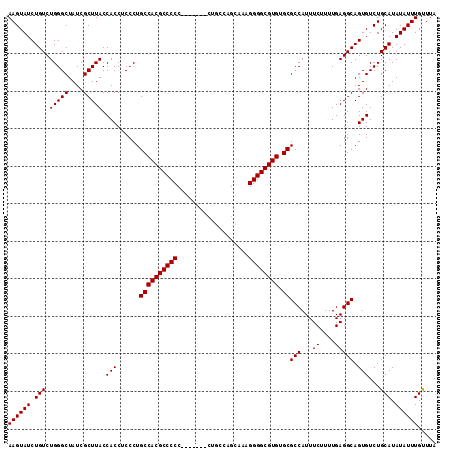
| Location | 13,213,640 – 13,213,754 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -31.78 |
| Energy contribution | -31.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13213640 114 - 23771897 UAAACAAAUAUAUGCAGACACUGCCUCAAAAGAAAUGGCGCACACGCCCCUUUGCUGGGCGUGGGGCAGGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUU ............(((.......(((...........)))...((((((((((.(((........))))))))))))))))(((..((.(....).))..)))............ ( -37.80) >DroSec_CAF1 6981 107 - 1 UAAACAAAUAUAUGCAGACACUGCCUCAAAAGAAAUGGCGCACACGCCCCUUUGCUGGCAG-------GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUU ............(((.......(((...........)))...((((((((((((....)))-------))))))))))))(((..((.(....).))..)))............ ( -38.70) >DroSim_CAF1 12054 107 - 1 UAAACAAAUAUAUGCAGACACUGCCUCAAAAGAAAUGGCGCACACGCCCCUUUGCUGGCUG-------GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUU ............(((.......(((...........)))...(((((((((..(....)..-------))))))))))))(((..((.(....).))..)))............ ( -35.30) >DroYak_CAF1 6890 105 - 1 UAAGCAAAUAUAUGCAGACACUGCCUCAAAAGAAAUGGCGCACACGCCCCU-UGCUGUCA--------GGGGGCGUGGCAGGGUGGUGGUAAGCGAUAGCCCAGACAGAUACUU ...(((......))).......(((...........)))((.(((((((((-((....))--------))))))))))).((((.((.(....).)).))))............ ( -39.00) >consensus UAAACAAAUAUAUGCAGACACUGCCUCAAAAGAAAUGGCGCACACGCCCCUUUGCUGGCAG_______GGGGGCGUGGCAGGGAGGUGGUAAGCGAUAGCCCAGACAGAUACUU ............(((.......(((...........)))...(((((((((.................))))))))))))(((..((.(....).))..)))............ (-31.78 = -31.78 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:14 2006