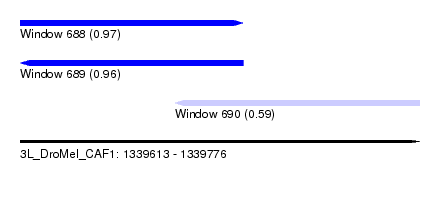
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,339,613 – 1,339,776 |
| Length | 163 |
| Max. P | 0.973173 |

| Location | 1,339,613 – 1,339,704 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.39 |
| Energy contribution | -29.45 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
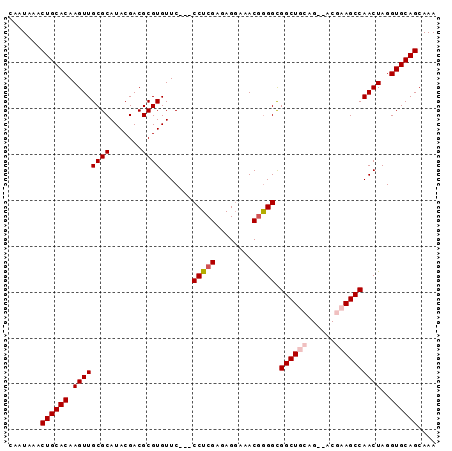
>3L_DroMel_CAF1 1339613 91 + 23771897 CAAUAAACUGCACAAGUUGCGCAUACGACGCGUGUUCCUCCCUCGAGAGGAAACGAGGUGGCUAAAG--ACGAAGCCAACUAGGUGCAGCAAA .......((((((..((((......)))).(((.((((((......)))))))))((.(((((....--....))))).))..)))))).... ( -29.10) >DroSec_CAF1 11842 88 + 1 CAAUAAACUGCACAAGUUGCGCAUACGACGCGUGUUC---CCUCGGGAGGAAACGGGGCGGCUGCAG--ACGCAGCCAACUAGGUGCAGCAAA .......((((((.((((((((.......))))....---(((((........))))).((((((..--..))))))))))..)))))).... ( -34.40) >DroSim_CAF1 11934 88 + 1 CAAUAAACUGCACAAGUUGCGCAUACGACGCGUGUUC---CCUCGGGAGGAAACGGGGCGGCUGCAG--ACGCAGCCAACUAGGUGCAGCAAA .......((((((.((((((((.......))))....---(((((........))))).((((((..--..))))))))))..)))))).... ( -34.40) >DroEre_CAF1 12196 90 + 1 CAAUAAACUGCACAAGUUGCGCAUACGACGCGUGUUC---CCUCGAGAGCAAACUGGGCGGCUGGGGGCGCGAAGCCAACUAGGUGCAGCAAA .......((((((.(((((.((........(((((((---((......((.......))....)))))))))..)))))))..)))))).... ( -30.80) >consensus CAAUAAACUGCACAAGUUGCGCAUACGACGCGUGUUC___CCUCGAGAGGAAACGGGGCGGCUGCAG__ACGAAGCCAACUAGGUGCAGCAAA .......((((((.((((((((.......)))).......(((((........))))).((((((......))))))))))..)))))).... (-28.39 = -29.45 + 1.06)

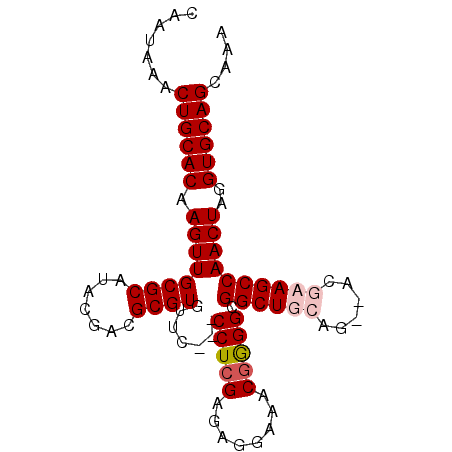
| Location | 1,339,613 – 1,339,704 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -27.91 |
| Energy contribution | -28.98 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1339613 91 - 23771897 UUUGCUGCACCUAGUUGGCUUCGU--CUUUAGCCACCUCGUUUCCUCUCGAGGGAGGAACACGCGUCGUAUGCGCAACUUGUGCAGUUUAUUG ...(((((((..((((((((....--....)))).....(((((((((....))))))).))((((.....)))))))).)))))))...... ( -33.80) >DroSec_CAF1 11842 88 - 1 UUUGCUGCACCUAGUUGGCUGCGU--CUGCAGCCGCCCCGUUUCCUCCCGAGG---GAACACGCGUCGUAUGCGCAACUUGUGCAGUUUAUUG ...(((((((..((((((((((..--..)))))).....(((((((....)))---))))..((((.....)))))))).)))))))...... ( -37.90) >DroSim_CAF1 11934 88 - 1 UUUGCUGCACCUAGUUGGCUGCGU--CUGCAGCCGCCCCGUUUCCUCCCGAGG---GAACACGCGUCGUAUGCGCAACUUGUGCAGUUUAUUG ...(((((((..((((((((((..--..)))))).....(((((((....)))---))))..((((.....)))))))).)))))))...... ( -37.90) >DroEre_CAF1 12196 90 - 1 UUUGCUGCACCUAGUUGGCUUCGCGCCCCCAGCCGCCCAGUUUGCUCUCGAGG---GAACACGCGUCGUAUGCGCAACUUGUGCAGUUUAUUG ...(((((((..(((((((..(((((((((((..((.......)).)).).))---).....))).))...)).))))).)))))))...... ( -27.80) >consensus UUUGCUGCACCUAGUUGGCUGCGU__CUGCAGCCGCCCCGUUUCCUCCCGAGG___GAACACGCGUCGUAUGCGCAACUUGUGCAGUUUAUUG ...(((((((..(((((((((((....))))))).....(((((((....)))...))))..((((.....)))))))).)))))))...... (-27.91 = -28.98 + 1.06)

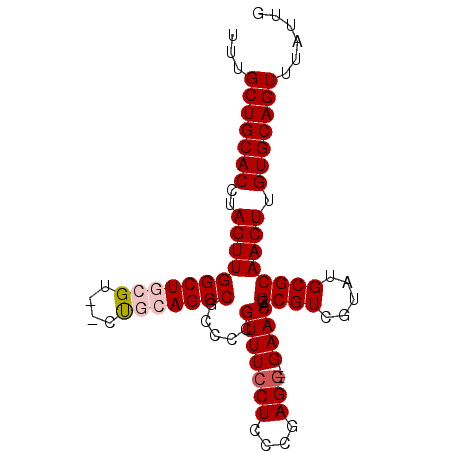
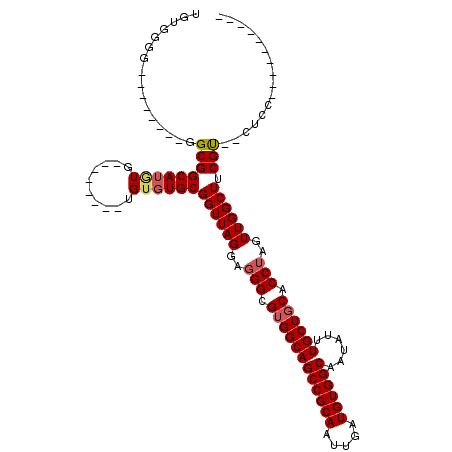
| Location | 1,339,676 – 1,339,776 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1339676 100 - 23771897 AGUGGGGGGAGGGGGGGCGGCAUGUG--------UGUGUGCGGUUAGAAGGGCGUGGCAGCCGCAAUUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUUCGU--CUUU---------- .......(((((.(((((.((((...--------...))))...(((..(((.(..(((((((((.....))))))......)))..).)))..)))))))).)--))))---------- ( -33.60) >DroSec_CAF1 11902 95 - 1 -GUGGGG--------GGCGGCAUGUGUGUG----UGUGUGCGGUUAGGAGGGCGUGGCAGCCGCAACUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUGCGU--CUGC---------- -...(.(--------(((.(((..(....)----..)))((((((((..(((.(..(((((((((.....))))))......)))..).)))..))))))))))--)).)---------- ( -36.20) >DroSim_CAF1 11994 100 - 1 UGUGGGG--------GGCGGCAUGUGAGUGUGUGUGAGUGCGGUUAGGAGGGCGUGGCAGCCGCAAUUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUGCGU--CUGC---------- ....(.(--------(((.(((..(....)..)))....((((((((..(((.(..(((((((((.....))))))......)))..).)))..))))))))))--)).)---------- ( -36.20) >DroEre_CAF1 12256 92 - 1 UGUGGGG--------GGCGGCAAGUG--------UGUGUGCGGUUAGG--GGCGGGGCAGCCGCAAUUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUUCGCGCCCCC---------- ...((((--------(((((((((((--------(.(((.(.((....--.)).).)))((((((.....)))))).))))))))))).((.....)).......)))))---------- ( -40.30) >DroYak_CAF1 12840 104 - 1 UGUGGGG--------GGCGGCAUAUG--------UGUGUGCGGUUAGGAGGGCGUGGCAGCCGCAAUUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUUCGCCCCCCUAACUUGCCAC ...((((--------(((((((((..--------..)))))((((((..(((.(..(((((((((.....))))))......)))..).)))..)))))).))))))))........... ( -47.60) >consensus UGUGGGG________GGCGGCAUGUG________UGUGUGCGGUUAGGAGGGCGUGGCAGCCGCAAUUGAUGUGGCAAUAUUUGCUGCACCUAGUUGGCUUCGU__CUCC__________ ................(((((((((..........))))))((((((..(((.((((((((((((.....))))))......)))))).)))..)))))).)))................ (-26.04 = -26.48 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:40 2006