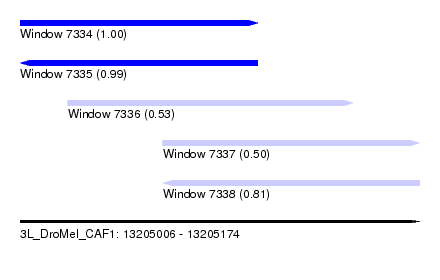
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,205,006 – 13,205,174 |
| Length | 168 |
| Max. P | 0.999831 |

| Location | 13,205,006 – 13,205,106 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -23.73 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13205006 100 + 23771897 ------GGAUGCUGCACUU--UGGCUGU------------UGCUGGUGUUUUGGUUCUUCCUUUUUAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCU ------...((((((.((.--.((((((------------.(((((......((.....))...))))).))....))))...)).))))))(((.((((((((....))))))))))). ( -29.00) >DroSim_CAF1 3627 101 + 1 ------AGAUGCUGCACUU--UGGCUGUU-----------UGCUGGUUUUUUGGUUCUUCCUUUUCAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCU ------...((((((.((.--.((((...-----------.(((((......((.....))...))))).......))))...)).))))))(((.((((((((....))))))))))). ( -31.10) >DroAna_CAF1 88802 116 + 1 GGGCCAAGAAGCUGCACUUCGUGGCUGCCACUUCCACUGCCACUGCCGCUGUUGUU----CUUUUCAGCUACUUAGAGCUUUUAGAGCGGCAGCAAGAUGUAAUAUAAAUUACAUUAGCU .(((...((((.....))))(((((.............))))).)))(((((((((----((....((((......))))...)))))))))))..((((((((....)))))))).... ( -41.52) >consensus ______AGAUGCUGCACUU__UGGCUGU____________UGCUGGUGUUUUGGUUCUUCCUUUUCAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCU ..........(((((......((((((..............((..(....)..))..........))))))......((((...)))).)))))..((((((((....)))))))).... (-23.73 = -22.85 + -0.88)

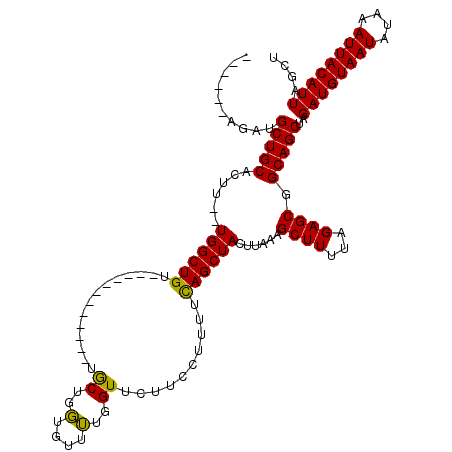
| Location | 13,205,006 – 13,205,106 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.79 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13205006 100 - 23771897 AGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAAAGCUUUAAGUAGCUAAAAAGGAAGAACCAAAACACCAGCA------------ACAGCCA--AAGUGCAGCAUCC------ ((((((((((((....))))))).)))))((.((.((...((((......)))).....((.....))............------------.......--.)).)).))....------ ( -20.70) >DroSim_CAF1 3627 101 - 1 AGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAAAGCUUUAAGUAGCUGAAAAGGAAGAACCAAAAAACCAGCA-----------AACAGCCA--AAGUGCAGCAUCU------ ((((((((((((....))))))).)))))((.((.((...((((......)))).....((.....))............-----------........--.)).)).))....------ ( -20.70) >DroAna_CAF1 88802 116 - 1 AGCUAAUGUAAUUUAUAUUACAUCUUGCUGCCGCUCUAAAAGCUCUAAGUAGCUGAAAAG----AACAACAGCGGCAGUGGCAGUGGAAGUGGCAGCCACGAAGUGCAGCUUCUUGGCCC .(((((((((((....))))))).(..((((((((((....((.....)).((((.....----.....)))))).))))))))..)...)))).((((.((((.....)))).)))).. ( -39.80) >consensus AGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAAAGCUUUAAGUAGCUGAAAAGGAAGAACCAAAACACCAGCA____________ACAGCCA__AAGUGCAGCAUCU______ ((((((((((((....))))))).)))))((.((.((...((((......)))).......................((................)).....)).)).)).......... (-16.68 = -16.79 + 0.11)

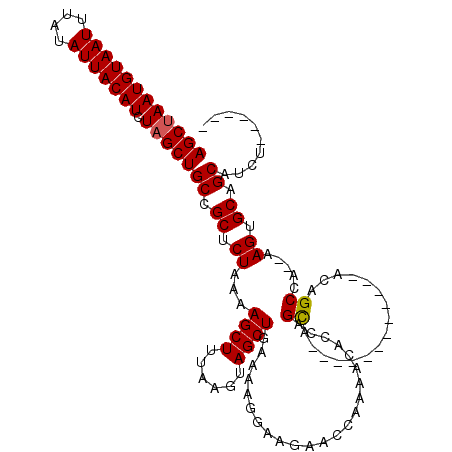
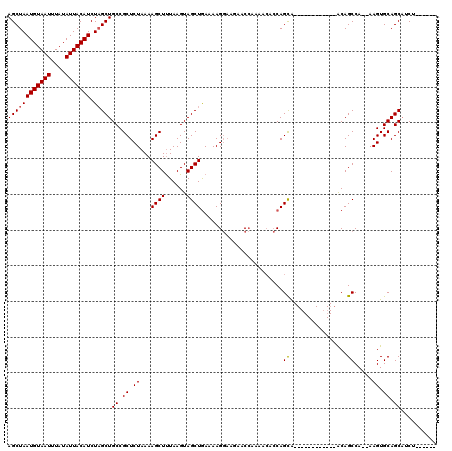
| Location | 13,205,026 – 13,205,146 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -21.90 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13205026 120 + 23771897 UGCUGGUGUUUUGGUUCUUCCUUUUUAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAA .((.((((((((((.....))......((..(((...((((...))))(((((((.((((((((....)))))))))))....)))))))..))............)))))))).))... ( -29.80) >DroSim_CAF1 3648 120 + 1 UGCUGGUUUUUUGGUUCUUCCUUUUCAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAA .....(((((((((((((........((((......))))...)))))...((((.((((((((....))))))))))))...((((....))))........))))))))......... ( -27.00) >DroAna_CAF1 88842 115 + 1 CACUGCCGCUGUUGUU----CUUUUCAGCUACUUAGAGCUUUUAGAGCGGCAGCAAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAA-AACAAAAAAUACUCGACAA ..((((((((((((((----((....((((......))))...)))))))))))..((((((((....)))))))).((.....))..))))).......-................... ( -35.20) >consensus UGCUGGUGUUUUGGUUCUUCCUUUUCAGCUACUUAAAGCUUUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAA .((..(....)..))............((..(((...((((...))))(((((((.((((((((....)))))))))))....)))))))..)).......................... (-21.90 = -21.57 + -0.33)

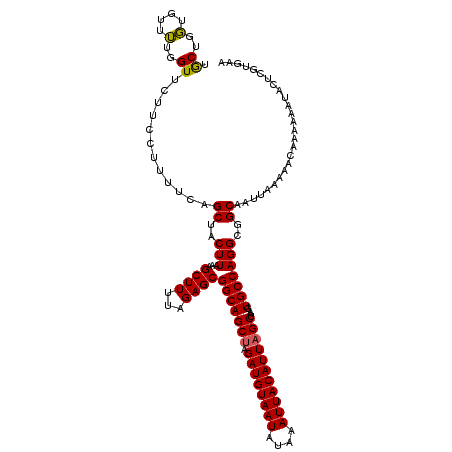
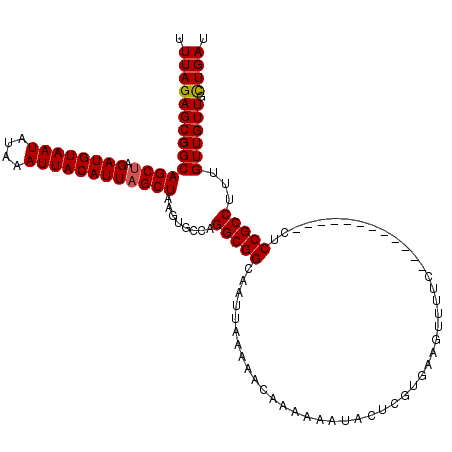
| Location | 13,205,066 – 13,205,174 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13205066 108 + 23771897 UUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAAGUUUUC------------CUCCGCCUUUGUUGUUGCUGAU .....(((((((((..((((((((....)))))))).((.....)).((((((......(((((................))))).------------..))))))..)))))))))... ( -27.29) >DroSim_CAF1 3688 108 + 1 UUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAAGUUUUC------------CUCCGCCUUUGUUGUUGCUGAU .....(((((((((..((((((((....)))))))).((.....)).((((((......(((((................))))).------------..))))))..)))))))))... ( -27.29) >DroAna_CAF1 88878 119 + 1 UUUAGAGCGGCAGCAAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAA-AACAAAAAAUACUCGACAACUUCUUCCUUCGGUGCCUCUCCGCCGUUGUUGUUCUUGAU ...(((((((((((..((((((((....))))))))...(((.((((....)))).))).-..............................((((......))))))))))))))).... ( -29.50) >consensus UUUAGAGCGGCAGCUAGAUGUAAUAUAAAUUACAUUAGCUAAGUGCCAGGCGGCAAUUAAAAACAAAAAAUACUCGUGAAGUUUUC____________CUCCGCCUUUGUUGUUGCUGAU .((((((((((((((.((((((((....))))))))))))........(((((...............................................)))))...)))))).)))). (-22.38 = -22.50 + 0.11)

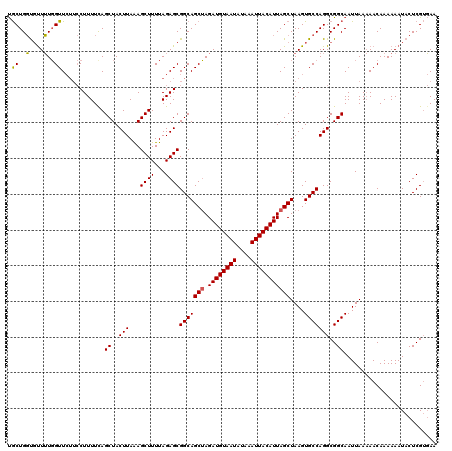
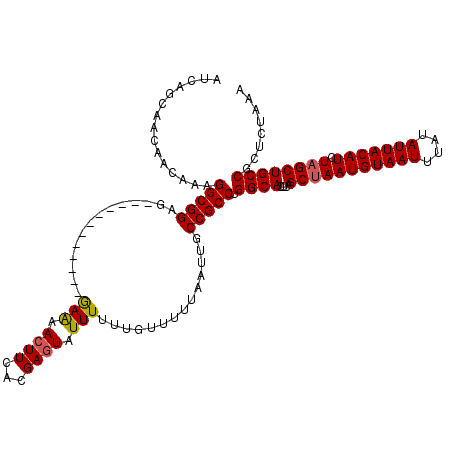
| Location | 13,205,066 – 13,205,174 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.39 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13205066 108 - 23771897 AUCAGCAACAACAAAGGCGGAG------------GAAAACUUCACGAGUAUUUUUUGUUUUUAAUUGCCGCCUGGCACUUAGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAA ...(((........((((((..------------((((((................)))))).....))))))((((....(((((((((((....))))))).)))))))))))..... ( -30.99) >DroSim_CAF1 3688 108 - 1 AUCAGCAACAACAAAGGCGGAG------------GAAAACUUCACGAGUAUUUUUUGUUUUUAAUUGCCGCCUGGCACUUAGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAA ...(((........((((((..------------((((((................)))))).....))))))((((....(((((((((((....))))))).)))))))))))..... ( -30.99) >DroAna_CAF1 88878 119 - 1 AUCAAGAACAACAACGGCGGAGAGGCACCGAAGGAAGAAGUUGUCGAGUAUUUUUUGUU-UUAAUUGCCGCCUGGCACUUAGCUAAUGUAAUUUAUAUUACAUCUUGCUGCCGCUCUAAA ..............((((((...((((..((((.(((((((........))))))).))-))...)))).))........(((..(((((((....)))))))...)))))))....... ( -28.10) >consensus AUCAGCAACAACAAAGGCGGAG____________GAAAACUUCACGAGUAUUUUUUGUUUUUAAUUGCCGCCUGGCACUUAGCUAAUGUAAUUUAUAUUACAUCUAGCUGCCGCUCUAAA ...............(((((..............(((.((((...)))).)))..............))))).((((....(((((((((((....))))))).))))))))........ (-22.16 = -22.39 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:03 2006