
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,193,820 – 13,193,912 |
| Length | 92 |
| Max. P | 0.972232 |

| Location | 13,193,820 – 13,193,912 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13193820 92 + 23771897 -GUUUUUAAG-----------------AGGUAUCUCUUAGGCGACCCUA-AGCCCUUUUUAAAUAAAAAGGCCUAGGGUGCUGUCAAUAGGAUUCCCUCGCAAAAGCGAAC--------- -((((((..(-----------------(((.((((....(((.((((((-.(((.(((((....)))))))).))))))))).......))))..))))..))))))....--------- ( -31.40) >DroSec_CAF1 45121 117 + 1 -GUUUUUAAGAUCGCUCAGCAUUUACUAGGUAUCUGUGAGGCGACCCCU-AGCCCUUU-AAGAAAAAAACGCCUAGGGUGCUGUCAAUAGGAUUCCCUCGCAAAAGCGAACCGCUGGCAC -............((.((((........((.((((.(((((((..((((-((.(.(((-.......))).).)))))))))).)))...)))).)).((((....))))...)))))).. ( -33.20) >DroSim_CAF1 43562 117 + 1 -CUUUUUAAGAUCUCCCAGCAUUUACUAGGUAUCUGUUUGGCGACCCCU-AGCCCUUU-AAGAAAAAAAGGCCUAGGGUGCUGUCAAUAGGAUUCCCUCGUAAAAGCGAACCGCUGGCAC -..............(((((........((.((((..((((((.(((((-((.(((((-.......))))).)))))).).))))))..)))).)).((((....))))...)))))... ( -38.30) >DroEre_CAF1 48757 97 + 1 GUUUCCAA-------------------AGGUAUCUGAUAGGCGACCCCAGCGCGCUCU----UGGAAAAGGCCUAGGGUGCUGUCAAUAGGAUUCCCUCGCAAAAGCGAACCGCUGGCAC ........-------------------.(((..((....))..)))((((((((((((----.((......)).)))))))........((....))((((....))))...)))))... ( -28.20) >consensus _GUUUUUAAG_________________AGGUAUCUGUUAGGCGACCCCA_AGCCCUUU_AAGAAAAAAAGGCCUAGGGUGCUGUCAAUAGGAUUCCCUCGCAAAAGCGAACCGCUGGCAC ............................((.((((....(((.((((....((.((((........))))))...))))))).......)))).)).((((....))))........... (-18.04 = -18.35 + 0.31)

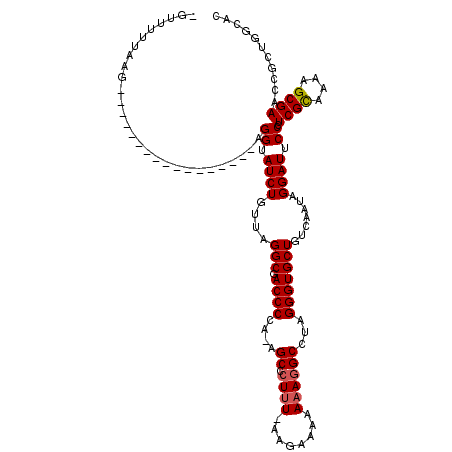

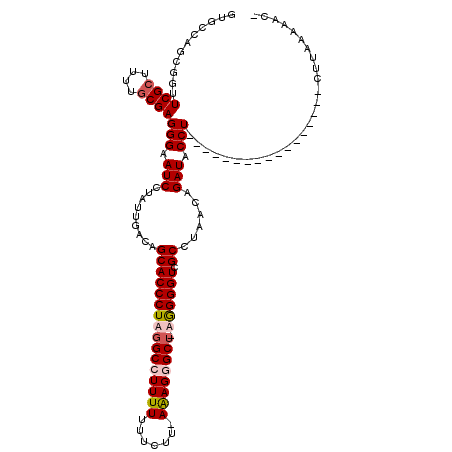
| Location | 13,193,820 – 13,193,912 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -19.93 |
| Energy contribution | -21.06 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13193820 92 - 23771897 ---------GUUCGCUUUUGCGAGGGAAUCCUAUUGACAGCACCCUAGGCCUUUUUAUUUAAAAAGGGCU-UAGGGUCGCCUAAGAGAUACCU-----------------CUUAAAAAC- ---------.(((((....)))))...............(((((((((((((((((.....)))))))))-)))))).)).((((((....))-----------------)))).....- ( -35.40) >DroSec_CAF1 45121 117 - 1 GUGCCAGCGGUUCGCUUUUGCGAGGGAAUCCUAUUGACAGCACCCUAGGCGUUUUUUUCUU-AAAGGGCU-AGGGGUCGCCUCACAGAUACCUAGUAAAUGCUGAGCGAUCUUAAAAAC- .((((((((..((((....))))(((.(((....(((..(((((((.(((.((((......-)))).)))-.))))).)).)))..))).)))......))))).)))...........- ( -36.20) >DroSim_CAF1 43562 117 - 1 GUGCCAGCGGUUCGCUUUUACGAGGGAAUCCUAUUGACAGCACCCUAGGCCUUUUUUUCUU-AAAGGGCU-AGGGGUCGCCAAACAGAUACCUAGUAAAUGCUGGGAGAUCUUAAAAAG- ((....((((...(((.(((..(((....)))..))).))).((((((.(((((.......-))))).))-)))).))))...))((((.((((((....))))))..)))).......- ( -38.40) >DroEre_CAF1 48757 97 - 1 GUGCCAGCGGUUCGCUUUUGCGAGGGAAUCCUAUUGACAGCACCCUAGGCCUUUUCCA----AGAGCGCGCUGGGGUCGCCUAUCAGAUACCU-------------------UUGGAAAC ...((((.(((((((....))))..........((((..(((((((((((((((....----)))).)).))))))).))...))))..))).-------------------)))).... ( -31.50) >consensus GUGCCAGCGGUUCGCUUUUGCGAGGGAAUCCUAUUGACAGCACCCUAGGCCUUUUUUUCUU_AAAGGGCU_AGGGGUCGCCUAACAGAUACCU_________________CUUAAAAAC_ ...........((((....))))(((.(((.........((((((((((((((((.......)))))))).)))))).))......))).)))........................... (-19.93 = -21.06 + 1.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:59 2006