| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,192,854 – 13,192,968 |
| Length | 114 |
| Max. P | 0.684997 |

| Location | 13,192,854 – 13,192,968 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192854 114 + 23771897 CUUAGGGUCUUGAACCACCUAACCUUUGUUACACAACCCU---GCCCUUCUGGGGUCACCGACCCUCGACGAACAAGUGUCGUUGCGUUGCACACAUGACACGAGAUCGUUUGGCCC ...(((((..(((((............))).))..)))))---(((.....((((((...)))))).(((((.(..((((((((((...)))...)))))))..).))))).))).. ( -32.40) >DroSec_CAF1 44129 107 + 1 CUGAGGGCCUUGACC--CCUAACCUUUGUUACACAACCCU---GCCCUUCUAGGGUCACCGACCCUCGACGAACAAGUGUCGU-----UGCACACAUGACACGAGAUCGUCUGGCCC ..((((((..(((((--(.((((....))))..((....)---)........))))))..).)))))(((((.(..(((((((-----.......)))))))..).)))))...... ( -33.20) >DroSim_CAF1 42557 107 + 1 CUUAGGGCCUUGACC--CCUAACCUUUGUUAUACAACCCU---GCCCUUCUGGGGUCACCGACCCUCGACGAACAAGUGUCGU-----UGCACACAUGACACGAGAUCGUUUGGCCC ....(((((.(((((--((.....................---........))))))).........(((((.(..(((((((-----.......)))))))..).))))).))))) ( -33.95) >DroEre_CAF1 47824 107 + 1 CCAAGGGCCUUGACC--CCUAACCUUUGUUACACACGCCG---CCCCCUCUGGGGUCGCCGACCCUUGACGAACAAGUGUCGU-----UGCACACAUGACACGAGAUCGUUGGGCCC ....((((((.....--...................((.(---((((....))))).))........(((((.(..(((((((-----.......)))))))..).))))))))))) ( -35.00) >DroYak_CAF1 43916 110 + 1 CCAAGGGCCUUGACC--CCUAACCUUUGCUACACACCCCUGCUGCCCUUCUGAGGUCACCGACCCUCGACGAACAAGUGUCGU-----UGCACACAUGACACGAGAUCGCUUGGCCC ....(((((.(((((--.(........((..((......))..))......).))))).........(.(((.(..(((((((-----.......)))))))..).))))..))))) ( -27.24) >consensus CUAAGGGCCUUGACC__CCUAACCUUUGUUACACAACCCU___GCCCUUCUGGGGUCACCGACCCUCGACGAACAAGUGUCGU_____UGCACACAUGACACGAGAUCGUUUGGCCC ....(((((..........................................((((((...)))))).(((((.(..(((((((............)))))))..).))))).))))) (-27.72 = -27.64 + -0.08)

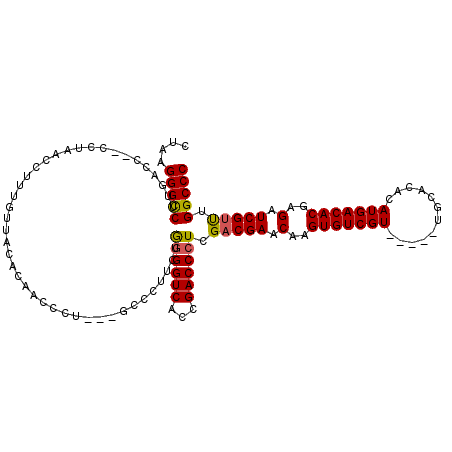

| Location | 13,192,854 – 13,192,968 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -30.15 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192854 114 - 23771897 GGGCCAAACGAUCUCGUGUCAUGUGUGCAACGCAACGACACUUGUUCGUCGAGGGUCGGUGACCCCAGAAGGGC---AGGGUUGUGUAACAAAGGUUAGGUGGUUCAAGACCCUAAG ((((((...(((((..(((..((((.....))))(((((.(((((((.((..(((((...)))))..)).))))---))))))))...))).)))))...))))))........... ( -38.00) >DroSec_CAF1 44129 107 - 1 GGGCCAGACGAUCUCGUGUCAUGUGUGCA-----ACGACACUUGUUCGUCGAGGGUCGGUGACCCUAGAAGGGC---AGGGUUGUGUAACAAAGGUUAGG--GGUCAAGGCCCUCAG (((((....((((((....(...(((...-----(((((.(((((((.((.((((((...)))))).)).))))---))))))))...)))...)...))--))))..))))).... ( -45.80) >DroSim_CAF1 42557 107 - 1 GGGCCAAACGAUCUCGUGUCAUGUGUGCA-----ACGACACUUGUUCGUCGAGGGUCGGUGACCCCAGAAGGGC---AGGGUUGUAUAACAAAGGUUAGG--GGUCAAGGCCCUAAG (((((....((((((....(...(((...-----(((((.(((((((.((..(((((...)))))..)).))))---))))))))...)))...)...))--))))..))))).... ( -43.50) >DroEre_CAF1 47824 107 - 1 GGGCCCAACGAUCUCGUGUCAUGUGUGCA-----ACGACACUUGUUCGUCAAGGGUCGGCGACCCCAGAGGGGG---CGGCGUGUGUAACAAAGGUUAGG--GGUCAAGGCCCUUGG (((((....((((((.(((.(((..(((.-----.((((.((((.....)))).)))).((.((((....))))---)))))..))).))).......))--))))..))))).... ( -42.90) >DroYak_CAF1 43916 110 - 1 GGGCCAAGCGAUCUCGUGUCAUGUGUGCA-----ACGACACUUGUUCGUCGAGGGUCGGUGACCUCAGAAGGGCAGCAGGGGUGUGUAGCAAAGGUUAGG--GGUCAAGGCCCUUGG (((((...((((((((((.((.((((...-----...)))).))..)).))).))))).(((((((.((...((.(((......))).)).....)).))--))))).))))).... ( -40.00) >consensus GGGCCAAACGAUCUCGUGUCAUGUGUGCA_____ACGACACUUGUUCGUCGAGGGUCGGUGACCCCAGAAGGGC___AGGGUUGUGUAACAAAGGUUAGG__GGUCAAGGCCCUAAG (((((..(((((((.(((((................)))))..((((.((..(((((...)))))..)).))))....))))))).((((....))))..........))))).... (-30.15 = -30.67 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:57 2006