
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,192,508 – 13,192,660 |
| Length | 152 |
| Max. P | 0.971744 |

| Location | 13,192,508 – 13,192,620 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.66 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192508 112 + 23771897 -----CGAAAUCUUGAAAUCCGUCGCUUGCCGCUUGGCGCUUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUUUUUUCAUAUGCUCCGCGCUCUAUUUU -----........(((((......((.....))((((((((.(((.((---.(((((((((.....))).)))))).)).)))))))))))....)))))...((.....))........ ( -32.40) >DroSec_CAF1 43774 111 + 1 -----CGAAAUCUUGAAAUCCGUCGCUUGCCGCUUGGCGCUUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUCCGCGCUCUAUUUC -----.(((((..(((((......((.....))((((((((.(((.((---.(((((((((.....))).)))))).)).)))))))))))...-)))))...((.....))...))))) ( -33.20) >DroSim_CAF1 42186 111 + 1 -----CGAAAUCUUGAAAUCCGUCGCUUGCCGCUUGGCGCUUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUCCGCGCUCUAUUUC -----.(((((..(((((......((.....))((((((((.(((.((---.(((((((((.....))).)))))).)).)))))))))))...-)))))...((.....))...))))) ( -33.20) >DroEre_CAF1 47459 104 + 1 -----CGAAAUCUCGAAAUCCGUCUUUU-------GGCGCUUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUGCGCGCUCUAUUUC -----.(((((...((((........((-------((((((.(((.((---.(((((((((.....))).)))))).)).)))))))))))...-))))....((.....))...))))) ( -31.10) >DroYak_CAF1 43525 112 + 1 AAUCUCGAAAUCUUGAAAUCCGUCGCUU-------GGCGCUUGACUCUUGUUGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGUUCCGCGCUCUAUUUC ......(((....(((((........((-------((((((.(((.((....(((((((((.....))).)))))).)).)))))))))))...-)))))....)))............. ( -28.80) >consensus _____CGAAAUCUUGAAAUCCGUCGCUUGCCGCUUGGCGCUUGACUCU___UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU_UUUCAUAUGCUCCGCGCUCUAUUUC ......(((((...((...................((((((.(((.((....(((((((((.....))).)))))).)).)))))))))..............((.....)))).))))) (-25.98 = -25.66 + -0.32)



| Location | 13,192,508 – 13,192,620 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -24.03 |
| Energy contribution | -25.43 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192508 112 - 23771897 AAAAUAGAGCGCGGAGCAUAUGAAAAAAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCA---AGAGUCAAGCGCCAAGCGGCAAGCGACGGAUUUCAAGAUUUCG----- ......((((.....))...(((((....(((((((((((.((..((.((.(((.....))))).))..---)).))).))))))))((.....))......))))).....)).----- ( -31.30) >DroSec_CAF1 43774 111 - 1 GAAAUAGAGCGCGGAGCAUAUGAAA-AAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCA---AGAGUCAAGCGCCAAGCGGCAAGCGACGGAUUUCAAGAUUUCG----- (((((...((.....))...(((((-...(((((((((((.((..((.((.(((.....))))).))..---)).))).))))))))((.....))......)))))..))))).----- ( -34.10) >DroSim_CAF1 42186 111 - 1 GAAAUAGAGCGCGGAGCAUAUGAAA-AAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCA---AGAGUCAAGCGCCAAGCGGCAAGCGACGGAUUUCAAGAUUUCG----- (((((...((.....))...(((((-...(((((((((((.((..((.((.(((.....))))).))..---)).))).))))))))((.....))......)))))..))))).----- ( -34.10) >DroEre_CAF1 47459 104 - 1 GAAAUAGAGCGCGCAGCAUAUGAAA-AAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCA---AGAGUCAAGCGCC-------AAAAGACGGAUUUCGAGAUUUCG----- (((((.((((.....))........-...(((((((((((.((..((.((.(((.....))))).))..---)).))).))))))-------))..........))...))))).----- ( -30.40) >DroYak_CAF1 43525 112 - 1 GAAAUAGAGCGCGGAACAUAUGAAA-AAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCAACAAGAGUCAAGCGCC-------AAGCGACGGAUUUCAAGAUUUCGAGAUU (((((....(((........(....-)..(((((((((((.((........(((((...)))))........)).))).))))))-------)))))....))))).............. ( -30.29) >consensus GAAAUAGAGCGCGGAGCAUAUGAAA_AAAUUGGCGCUGACACUGACAAGUCGUGUGAAGCACACCUGCA___AGAGUCAAGCGCCAAGCGGCAAGCGACGGAUUUCAAGAUUUCG_____ (((((....(((....(....).......(((((((((((.((........(((((...)))))........)).))).)))))))).......)))....))))).............. (-24.03 = -25.43 + 1.40)

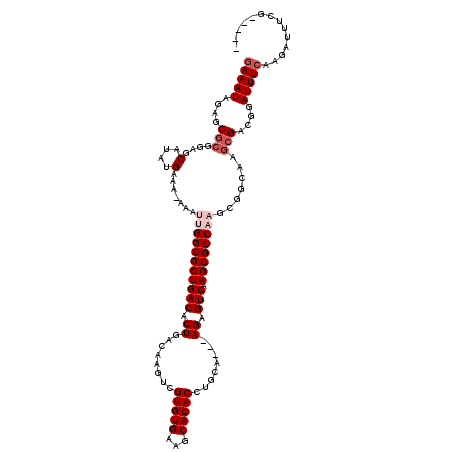

| Location | 13,192,543 – 13,192,660 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -27.87 |
| Energy contribution | -28.15 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192543 117 + 23771897 UUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUUUUUUCAUAUGCUCCGCGCUCUAUUUUUUCGAGUUUUGUGGCAUAGACAAAGCCAUGGCAGGCAUUU (((((.((---.(((((((((.....))).)))))).)).))))).(((....(((.((.(((((..((.((((.........))))..))..))))))).)))((....)).))).... ( -34.70) >DroSec_CAF1 43809 115 + 1 UUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUCCGCGCUCUAUUUCUUCGAGUUUUGUGGCAUAGACAAAG-CAUGGCAGACAUUU (((((.((---.(((((((((.....))).)))))).)).))))).((((.(((-(.((.(((((..((.((((.........))))..))..))))))).))))-..))))........ ( -33.40) >DroSim_CAF1 42221 115 + 1 UUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUCCGCGCUCUAUUUCUUCGAGUUUUGUGGCAUAGACAAAG-CAUGGCAGACAUUU (((((.((---.(((((((((.....))).)))))).)).))))).((((.(((-(.((.(((((..((.((((.........))))..))..))))))).))))-..))))........ ( -33.40) >DroEre_CAF1 47487 115 + 1 UUGACUCU---UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGCUGCGCGCUCUAUUUCUUCGAGUUUUGUGGCUUAGACAAAG-CAUGGCAGGCAUUU .((((.((---.(((((((((.....))).)))))).)).))))((((((.(((-(.((.((.((..((.((((.........))))..))..)).)))).))))-..))))..)).... ( -32.30) >DroYak_CAF1 43558 118 + 1 UUGACUCUUGUUGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU-UUUCAUAUGUUCCGCGCUCUAUUUCUUUGAGUUUUGUGGCAUGGACAAAG-CAAAGCAGGCAUUU ......((((((((..(((((...)))))...(((((.(((((((((...((..-......))....)))((((.........))))....)))))).))))).)-))..)))))..... ( -28.00) >consensus UUGACUCU___UGCAGGUGUGCUUCACACGACUUGUCAGUGUCAGCGCCAAUUU_UUUCAUAUGCUCCGCGCUCUAUUUCUUCGAGUUUUGUGGCAUAGACAAAG_CAUGGCAGGCAUUU (((((.((....(((((((((.....))).)))))).)).))))).((((..........(((((..((.((((.........))))..))..)))))..........))))........ (-27.87 = -28.15 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:55 2006