
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,192,265 – 13,192,453 |
| Length | 188 |
| Max. P | 0.989035 |

| Location | 13,192,265 – 13,192,377 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13192265 112 - 23771897 CUUAAAACUGGUGCAACACACAUUGGCCUCCUCCUCCGCCGACUCGAACAAAAAAAAUCCCCCGACAUGUGUA-----C---CUCCUGCCCUGAAACCUGGCAGAGCGCCAAACUAGCCG ........((((((...((((((((((..........))))).(((................)))..))))).-----.---...(((((..(....).))))).))))))......... ( -25.69) >DroSec_CAF1 43538 108 - 1 CUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCCAACUCGAA--AACAAAAUCCCCCGACAUGUGUA-----CCACCUCCUGUCCUGAAACCUGGCAGAGCGCCAAACUAGCCG ........((((((.(((--(((((((...---....))))).(((..--............)))..))))).-----.......(((.((........))))).))))))......... ( -24.74) >DroSim_CAF1 41952 106 - 1 CUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCCGACUCGAA--A--AAAAUCCCCCGACAUGUGUA-----CCACCUCCUGCCCUGAAACCUGGCAGAGCGCCAAACUAGCCG ........((((((.(((--(((((((...---....))))).(((..--.--.........)))..))))).-----.......(((((..(....).))))).))))))......... ( -26.60) >DroEre_CAF1 47226 102 - 1 CUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCCGACUCGAA-----AAAAUCCCCCGACAUGUGU--------ACCUCCUGCCCUGAAAGCUGGCAAAGCGCCAAACUAGCCG ........((((((.(((--(((((((...---....))))).(((..-----.........)))..)))))--------......((((.........))))..))))))......... ( -23.40) >DroYak_CAF1 43262 110 - 1 CUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCCGACUCGAA-----AAAAUCCCCCGACAUGUGUAUGUUCCUACCUCCUUCCCUGGAAGCUGGCAGAGCGCCAAACUAGCCG ........((((((.(((--(((((((...---....))))).(((..-----.........)))..)))))......((.((..((((....))))..)).)).))))))......... ( -27.90) >consensus CUUAAAACUGGUGCAACA__CAUUGGCCUC___CUCCGCCGACUCGAA__A__AAAAUCCCCCGACAUGUGUA_____C_ACCUCCUGCCCUGAAACCUGGCAGAGCGCCAAACUAGCCG ........((((((........(((((..........)))))...........................................(((((..(....).))))).))))))......... (-19.18 = -19.26 + 0.08)

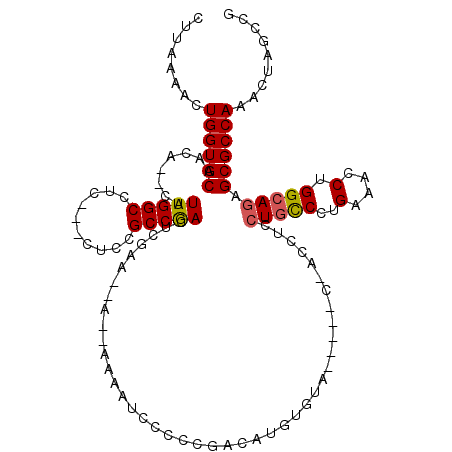
| Location | 13,192,337 – 13,192,453 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -35.34 |
| Energy contribution | -35.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
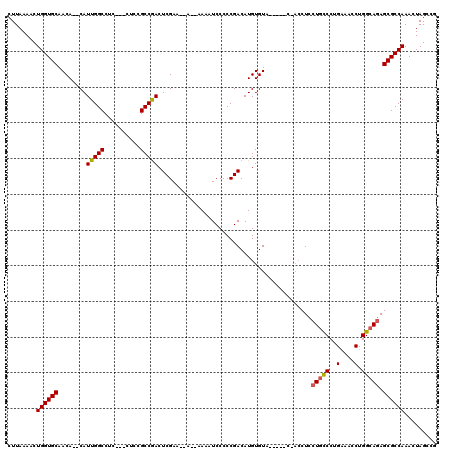
>3L_DroMel_CAF1 13192337 116 + 23771897 GGCGGAGGAGGAGGCCAAUGUGUGUUGCACCAGUUUUAAGGCCACGA----CGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((..((((.(((((.(((((((((..((........))...)))----))))))..)).)))((((..((((......(((((....)))))))))))))....))))...)))).. ( -43.00) >DroSec_CAF1 43611 111 + 1 GGCGGAG---GAGGCCAAUG--UGUUGCACCAGUUUUAAGGCCACGA----CGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((..---(((((.((((--(((((((.((((((....((.....----.))......)))))).....)))))))))))((((....))))............)))))...)))).. ( -38.60) >DroSim_CAF1 42023 111 + 1 GGCGGAG---GAGGCCAAUG--UGUUGCACCAGUUUUAAGGCCACGA----CGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((..---(((((.((((--(((((((.((((((....((.....----.))......)))))).....)))))))))))((((....))))............)))))...)))).. ( -38.60) >DroEre_CAF1 47293 115 + 1 GGCGGAG---GAGGCCAAUG--UGUUGCACCAGUUUUAAGGCCACGACGGACGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((..---(((((....(--(((((((((........))...((..((((((..(((...(.(((.....))).)..)))...))))))))..))))))))...)))))...)))).. ( -40.30) >DroYak_CAF1 43337 111 + 1 GGCGGAG---GAGGCCAAUG--UGUUGCACCAGUUUUAAGGCCACGA----CGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((..---(((((.((((--(((((((.((((((....((.....----.))......)))))).....)))))))))))((((....))))............)))))...)))).. ( -38.60) >consensus GGCGGAG___GAGGCCAAUG__UGUUGCACCAGUUUUAAGGCCACGA____CGCACAGCUGAGCUGCAUUUUGCAACAUGUUUCGGUGUCCCGAAUGCAAUGCCAUGCCUCAAGCCGCAA .((((.....(((((.......(((((((.((((((...(((..((.....))....))))))))).....)))))))((((((((....))))).))).......)))))...)))).. (-35.34 = -35.34 + -0.00)

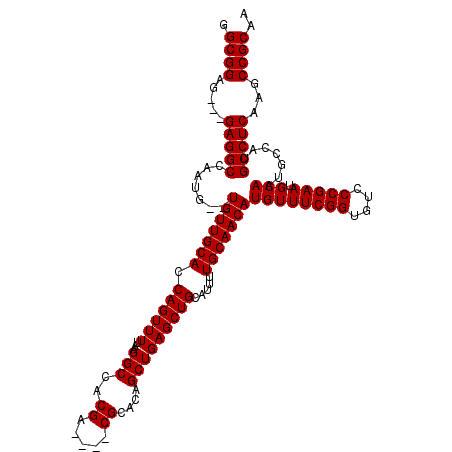
| Location | 13,192,337 – 13,192,453 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -38.90 |
| Energy contribution | -38.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
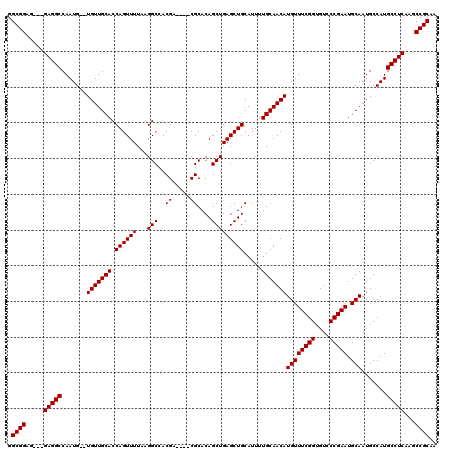
>3L_DroMel_CAF1 13192337 116 - 23771897 UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCG----UCGUGGCCUUAAAACUGGUGCAACACACAUUGGCCUCCUCCUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).))))(----(.((((((........)))....)))))....))))).....)))). ( -40.40) >DroSec_CAF1 43611 111 - 1 UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCG----UCGUGGCCUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).)))).----..((((((........)))....))--)....)))))---..)))). ( -40.90) >DroSim_CAF1 42023 111 - 1 UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCG----UCGUGGCCUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).)))).----..((((((........)))....))--)....)))))---..)))). ( -40.90) >DroEre_CAF1 47293 115 - 1 UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCGUCCGUCGUGGCCUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).)))).......((((((........)))....))--)....)))))---..)))). ( -40.90) >DroYak_CAF1 43337 111 - 1 UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCG----UCGUGGCCUUAAAACUGGUGCAACA--CAUUGGCCUC---CUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).)))).----..((((((........)))....))--)....)))))---..)))). ( -40.90) >consensus UUGCGGCUUGAGGCAUGGCAUUGCAUUCGGGACACCGAAACAUGUUGCAAAAUGCAGCUCAGCUGUGCG____UCGUGGCCUUAAAACUGGUGCAACA__CAUUGGCCUC___CUCCGCC ..((((...(((((...((((.((.(((((....)))))....(((((.....)))))...)).)))).......((.(((........)))...))........))))).....)))). (-38.90 = -38.90 + -0.00)

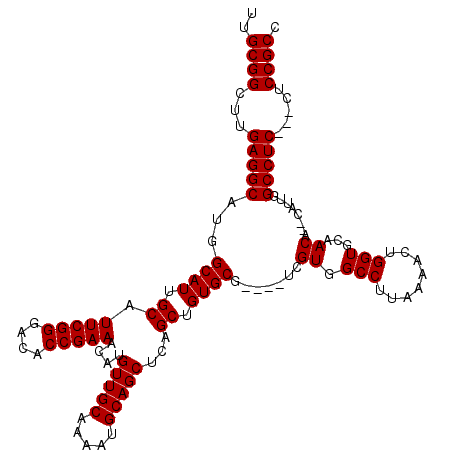
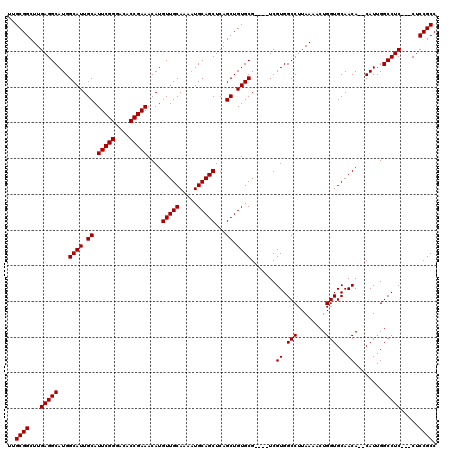
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:52 2006