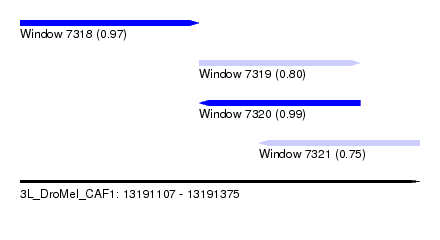
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,191,107 – 13,191,375 |
| Length | 268 |
| Max. P | 0.987752 |

| Location | 13,191,107 – 13,191,227 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13191107 120 + 23771897 CUCCAAUCCUCAAUCCUUGUCAGACUUUUCAGAGGAGGCGGCGAUGGGAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGGGACAAAGCCCGAAAACCGAAUUGAAAACCAA .........(((((((((....((....)).)))))((((((((((.......(((((.....))))).))))))).((((((....))))))..)))...........))))....... ( -33.60) >DroSec_CAF1 42404 113 + 1 CCCCAAU-------CCUCCUCAGACUUUUCAGAGAAGGCGGCGAUGGAAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGGGACAAAGCCCGAAAACCGAAUUGAAAACCAA .......-------...........(((((((....((((((((((.......(((((.....))))).))))))).((((((....))))))..)))((.....))..))))))).... ( -31.80) >DroSim_CAF1 40801 113 + 1 CCCCAAU-------CCUCGUCAGACUUUUCAGAGAAGGCGGCGAUGGGAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGGGACAAAGCCCGAAAACCGAAUUGAAAACCAA .((((.(-------(.(((((...(((....)))..))))).))))))...(((.......(((((......)))))((((((....))))))..)))...................... ( -31.80) >DroEre_CAF1 46051 112 + 1 -CCCAAU-------CCUCGAAAAACUUUUCAGGGCAGGCGGCGAUGGGAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGGGACAAAGCCCCAAAACCGAAUUGAAUGCCAA -((((..-------(((.((((....)))))))((.....))..))))...(((((.(((((((((......)))).((((((....))))))..............))))).))))).. ( -35.80) >DroYak_CAF1 42003 113 + 1 ACCCAAU-------CGUCGAAAAACUUUUCAGAGAAGGCGGCGAUGGGAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGUGACAAAGCCCGAAAACCGAAUUGAAAGCCAA .((((.(-------(((((.....((((.....)))).))))))))))...(((...(((((((((......))))...(((..(((.......)))..))).....)))))...))).. ( -34.60) >consensus CCCCAAU_______CCUCGUCAGACUUUUCAGAGAAGGCGGCGAUGGGAAAGGCAUGCAAUUGGCAUGACAUUGCCAUGUUCCAGCUGGGACAAAGCCCGAAAACCGAAUUGAAAACCAA .........................(((((((....((((((((((.......(((((.....))))).))))))).((((((....))))))..)))((.....))..))))))).... (-29.28 = -29.64 + 0.36)


| Location | 13,191,227 – 13,191,335 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -33.84 |
| Energy contribution | -35.84 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13191227 108 + 23771897 GGAGUGGGACUGCAAGCUAGUGCUGAUGGAAUAGCUCAGCCAUGGCGUUGGGGUCUG------------AGUUGGAGUCGCCACCGGUGUGGCAUGUGGAUGCCAUGUUGAUGCCGGCAC .((.(.(((((.(((((((..(((((.(......))))))..)))).))).))))).------------).))...((((.((.(((..((((((....))))))..))).)).)))).. ( -42.80) >DroSec_CAF1 42517 97 + 1 GGAGUGGGACUGCAAGCUAGUGCUGAUGGAAUAGUUCAGCCAUGGCGUUGGGGUCUG------------AGUCGGAGUCGCCACCGGUGUGGCAUGUGGAU-----------GCCGGCAC ....(.(((((.(((((((..(((((.........)))))..)))).))).))))).------------)(((((.((((((((....))))).....)))-----------.))))).. ( -37.90) >DroSim_CAF1 40914 108 + 1 GGAGUGGGACUGCAAGCUAGUGCUGAUGGAAUAGUUCAGCCAUGGCGUUGGGGUCUG------------AGUUGGAGUCGCCACCGGUGUGGCAUGUGGAUGCCAUGUUGAUGCCGGCAC .((.(.(((((.(((((((..(((((.........)))))..)))).))).))))).------------).))...((((.((.(((..((((((....))))))..))).)).)))).. ( -43.20) >DroEre_CAF1 46163 101 + 1 G-------GCUGCAAGCUAUUGCUGAUGGAAUAGUUCAGCCAUGGCGUUGGGGUCUG------------AGUCGGAGUCGCCACCGGUGUGGCAUGUGGAUGCCAUGUUGAUGCCGGCAC (-------(((.((((((((.(((((.........))))).))))).))).))))..------------.(((((.((((((((....)))))((((((...)))))).))).))))).. ( -42.50) >DroYak_CAF1 42116 120 + 1 GGAGUGGGGCUGCAAGCUAGUGCUGAUGGAAUAGUUCAGCCAUGGCGUUGGGAUCUGACUCGGAGUUGGAGUCGGAGUCGCCACCGGUGUGGCAUGUGGAUGCCAUGUUGAUGCCAGCAC ...(((.(((((..(((((.(.(....).).)))))))))).(((((((((((((((((((.......)))))))).)).))).(((..((((((....))))))..))))))))).))) ( -48.50) >consensus GGAGUGGGACUGCAAGCUAGUGCUGAUGGAAUAGUUCAGCCAUGGCGUUGGGGUCUG____________AGUCGGAGUCGCCACCGGUGUGGCAUGUGGAUGCCAUGUUGAUGCCGGCAC ......(((((.(((((((..(((((.........)))))..)))).))).)))))..............(((((.((((((((....)))))(((((.....))))).))).))))).. (-33.84 = -35.84 + 2.00)


| Location | 13,191,227 – 13,191,335 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -26.36 |
| Energy contribution | -27.56 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13191227 108 - 23771897 GUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCAACU------------CAGACCCCAACGCCAUGGCUGAGCUAUUCCAUCAGCACUAGCUUGCAGUCCCACUCC .(((.(((.....((((((........(((((....)))))((.......)------------)..........))))))(((((.........)))))....))).))).......... ( -32.50) >DroSec_CAF1 42517 97 - 1 GUGCCGGC-----------AUCCACAUGCCACACCGGUGGCGACUCCGACU------------CAGACCCCAACGCCAUGGCUGAACUAUUCCAUCAGCACUAGCUUGCAGUCCCACUCC (((..(((-----------((....))))).))).(((((.((((.(((((------------.((....((......))(((((.........))))).)))).))).))))))))).. ( -26.50) >DroSim_CAF1 40914 108 - 1 GUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCAACU------------CAGACCCCAACGCCAUGGCUGAACUAUUCCAUCAGCACUAGCUUGCAGUCCCACUCC .(((.(((.....((((((........(((((....)))))((.......)------------)..........))))))(((((.........)))))....))).))).......... ( -31.70) >DroEre_CAF1 46163 101 - 1 GUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACU------------CAGACCCCAACGCCAUGGCUGAACUAUUCCAUCAGCAAUAGCUUGCAGC-------C .(((.(((.....((((((........(((((....)))))((.......)------------)..........))))))(((((.........)))))....))).)))..-------. ( -31.70) >DroYak_CAF1 42116 120 - 1 GUGCUGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACUCCAACUCCGAGUCAGAUCCCAACGCCAUGGCUGAACUAUUCCAUCAGCACUAGCUUGCAGCCCCACUCC .(((.(((.....((((((........(((((....)))))(((((.((.......)).)))))..........))))))(((((.........)))))....))).))).......... ( -40.40) >consensus GUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACU____________CAGACCCCAACGCCAUGGCUGAACUAUUCCAUCAGCACUAGCUUGCAGUCCCACUCC .(((.(((.....((((((.......((((((....)))))).......................(....)...))))))(((((.........)))))....))).))).......... (-26.36 = -27.56 + 1.20)


| Location | 13,191,267 – 13,191,375 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13191267 108 - 23771897 GUUCCCUCGACUGCCUCGAAGUGCGUAAUGCGCUCAUUACGUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCAACU------------CAGACCCCAACGCCAUG ........((.((((.....(..(((((((....)))))))..).))))))..((((((........(((((....)))))((.......)------------)..........)))))) ( -33.40) >DroSec_CAF1 42557 97 - 1 GUCCCCUCGACUGCCUCGGAGUGCGUAAUGCGCUCAUUACGUGCCGGC-----------AUCCACAUGCCACACCGGUGGCGACUCCGACU------------CAGACCCCAACGCCAUG ..........(((..(((((((((((((((....))))))))((((((-----------((....)))))((....))))).)))))))..------------))).............. ( -33.20) >DroSim_CAF1 40954 108 - 1 GUCCCCUCGACUGCCUCGAAGUGCGUAAUGCGCUCAUUACGUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCAACU------------CAGACCCCAACGCCAUG ........((.((((.....(..(((((((....)))))))..).))))))..((((((........(((((....)))))((.......)------------)..........)))))) ( -33.40) >DroEre_CAF1 46196 108 - 1 GUCCCCUCGAAUGCCUCGAAGUGCGUAAUGCGCUCAUUACGUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACU------------CAGACCCCAACGCCAUG ........((.((((.....(..(((((((....)))))))..).))))))..((((((........(((((....)))))((.......)------------)..........)))))) ( -33.40) >DroYak_CAF1 42156 120 - 1 GUCCCCUCGACCGCCUCGAAGUGCGUAAUGCGCUCAUUACGUGCUGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACUCCAACUCCGAGUCAGAUCCCAACGCCAUG (((.....))).(((....((..(((((((....)))))))..))))).....((((((........(((((....)))))(((((.((.......)).)))))..........)))))) ( -43.00) >consensus GUCCCCUCGACUGCCUCGAAGUGCGUAAUGCGCUCAUUACGUGCCGGCAUCAACAUGGCAUCCACAUGCCACACCGGUGGCGACUCCGACU____________CAGACCCCAACGCCAUG (((...((((.....)))).(..(((((((....)))))))..).))).....((((((.......((((((....)))))).......................(....)...)))))) (-26.84 = -27.72 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:47 2006