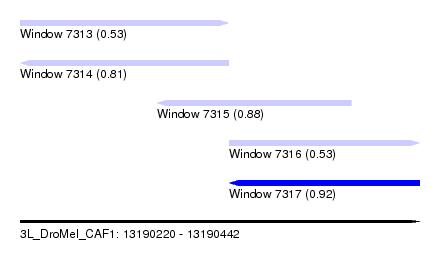
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,190,220 – 13,190,442 |
| Length | 222 |
| Max. P | 0.917777 |

| Location | 13,190,220 – 13,190,336 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -35.79 |
| Consensus MFE | -23.18 |
| Energy contribution | -24.98 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13190220 116 + 23771897 GUCUAAAAGCAUCACAGGGCCAAAAACACCUGCCUCGCACCUCGAUUCGAAG-GCCAUGAUA---UAAAGAGGGUUGUCAGGUGAGAGGAGGAGGUGCAGUAUUGGACCCUGUUGCAGUU ........(((..((((((((((.(.(((((.((((.(((((.(((.....(-(((......---.......)))))))))))).))))...)))))...).)))).))))))))).... ( -41.52) >DroSec_CAF1 41572 115 + 1 -UCUAAAAACAUCACAGGGCCAAAAACACCUGCCUCGCACCUCAAUUUGAAGGGCCAUGAUA---UAAAGAGGGUUGUCAGGUGAGAG-AGGAGGUGCAGUGUUGGACCCUGUUGCAGUU -............((((((((((...(((((.((((....((((.(((((..((((......---.......)))).))))))))).)-)))))))).....)))).))))))....... ( -38.22) >DroSim_CAF1 39947 114 + 1 -UCUAAAAACAUCACUGGGCCAAAAACACCUGCCUCGCACCUCAAUUUGAAG-GCCAUGAUA---UAAAGAGGGAUGUCGGAUGAGAG-AGGAGGUGCUGUGUUGGACCCUGUUGCAGUU -............((.(((((((...(((((.((((....((((.(((((..-.((......---.......))...))))))))).)-)))))))).....)))).))).))....... ( -32.12) >DroEre_CAF1 45214 113 + 1 GUCUAAAAACAUCACAGGGCCAAAAACACCUGCCUCGCACCUCCAUUCGAAG-GCCAUGAUA---UGGAGAGGACCAACAGGUGAG---AGGAGGUGAGUUGGUGGACCCUGUUGCAGUC .............(((((((((..(((((((.((((.((((((((((((...-....))).)---))))..(......).)))).)---)))))))..)))..))).))))))....... ( -38.70) >DroYak_CAF1 41102 106 + 1 GUCGAAAAACAUCACAGGGCCAAAAACACCUGCCUCGCACCUCAAUUUGAAG-GCCAUGAUACUACAAAGUGGAAUAACAGGUGAG---AGGAGGUG----------CUCUGCUGCAGUU .......(((..((((((((.......((((.((((.(((((...((((.((-.........)).))))((......))))))).)---))))))))----------))))).))..))) ( -28.41) >consensus GUCUAAAAACAUCACAGGGCCAAAAACACCUGCCUCGCACCUCAAUUUGAAG_GCCAUGAUA___UAAAGAGGGAUGUCAGGUGAGAG_AGGAGGUGCAGUGUUGGACCCUGUUGCAGUU .............((((((((((...(((((.(((.....(((.((((((....((...............))....)))))))))...)))))))).....)))).))))))....... (-23.18 = -24.98 + 1.80)

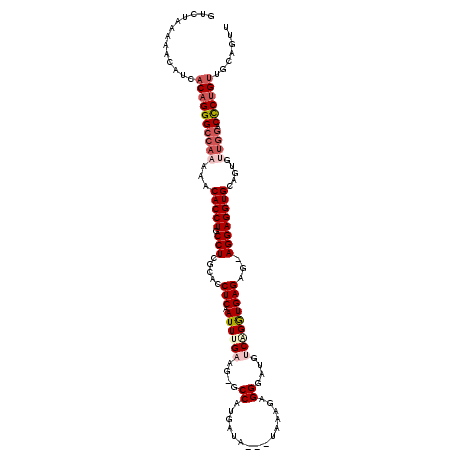
| Location | 13,190,220 – 13,190,336 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -23.98 |
| Energy contribution | -25.42 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13190220 116 - 23771897 AACUGCAACAGGGUCCAAUACUGCACCUCCUCCUCUCACCUGACAACCCUCUUUA---UAUCAUGGC-CUUCGAAUCGAGGUGCGAGGCAGGUGUUUUUGGCCCUGUGAUGCUUUUAGAC ....(((((((((.((((....((((((((((.((......))............---.......((-(((((...))))).)))))).))))))..))))))))))..)))........ ( -39.40) >DroSec_CAF1 41572 115 - 1 AACUGCAACAGGGUCCAACACUGCACCUCCU-CUCUCACCUGACAACCCUCUUUA---UAUCAUGGCCCUUCAAAUUGAGGUGCGAGGCAGGUGUUUUUGGCCCUGUGAUGUUUUUAGA- (((....((((((.((((....(((((((((-(((......))............---.......((((((......)))).)))))).))))))..))))))))))...)))......- ( -36.80) >DroSim_CAF1 39947 114 - 1 AACUGCAACAGGGUCCAACACAGCACCUCCU-CUCUCAUCCGACAUCCCUCUUUA---UAUCAUGGC-CUUCAAAUUGAGGUGCGAGGCAGGUGUUUUUGGCCCAGUGAUGUUUUUAGA- (((....((.(((.((((...((((((((((-(........((......))....---.......((-((((.....)))).)))))).))))))).))))))).))...)))......- ( -31.30) >DroEre_CAF1 45214 113 - 1 GACUGCAACAGGGUCCACCAACUCACCUCCU---CUCACCUGUUGGUCCUCUCCA---UAUCAUGGC-CUUCGAAUGGAGGUGCGAGGCAGGUGUUUUUGGCCCUGUGAUGUUUUUAGAC (((....((((((.(((......((((((((---(.(((((..(((......)))---........(-(.......))))))).)))).)))))....)))))))))...)))....... ( -40.30) >DroYak_CAF1 41102 106 - 1 AACUGCAGCAGAG----------CACCUCCU---CUCACCUGUUAUUCCACUUUGUAGUAUCAUGGC-CUUCAAAUUGAGGUGCGAGGCAGGUGUUUUUGGCCCUGUGAUGUUUUUCGAC ....((((.((((----------((((((((---(.((((((((((...(((....)))...)))))-..........))))).)))).))))))))).....))))............. ( -29.00) >consensus AACUGCAACAGGGUCCAACACUGCACCUCCU_CUCUCACCUGACAACCCUCUUUA___UAUCAUGGC_CUUCAAAUUGAGGUGCGAGGCAGGUGUUUUUGGCCCUGUGAUGUUUUUAGAC ....(((((((((.((((....(((((((((..........((......))..............((.((((.....)))).)).))).))))))..))))))))))..)))........ (-23.98 = -25.42 + 1.44)

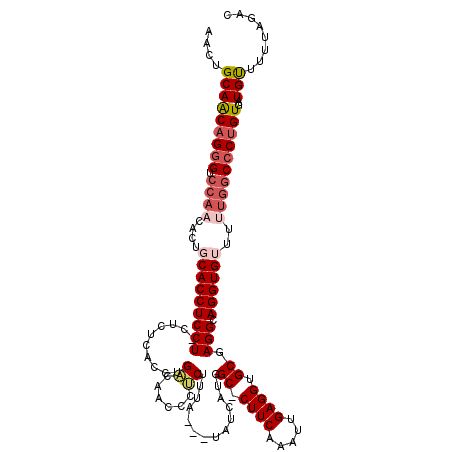
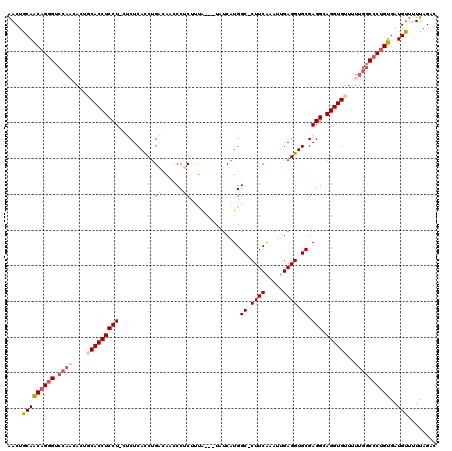
| Location | 13,190,296 – 13,190,404 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.47 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13190296 108 - 23771897 AGUGCAACAACAUCGGCAACAGCAGAGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCGAACUGCAACAGGGUCCAAUACUGCACCUCCUCCUCUCACC .(((((........(....).((((.((((((.......))))))((((.....))))............))))...............))))).............. ( -23.40) >DroSec_CAF1 41648 107 - 1 AGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACUCCGAACUGCAACAGGGUCCAACACUGCACCUCCU-CUCUCACC .(((((........(....).((((.((((((.......))))))((((.....))))............))))...............))))).....-........ ( -22.80) >DroSim_CAF1 40022 107 - 1 AGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACUCCGAACUGCAACAGGGUCCAACACAGCACCUCCU-CUCUCAUC .((((.........(....).((((.((((((.......))))))((((.....))))............))))................)))).....-........ ( -22.20) >DroEre_CAF1 45290 95 - 1 ----------CAUCGGCAACAGCAGCGCAUCAAAGUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCGGACUGCAACAGGGUCCACCAACUCACCUCCU---CUCACC ----------....((.....(((..((((((.......))))))..))).................(((((.......))))).))............---...... ( -19.20) >DroYak_CAF1 41181 95 - 1 AGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAGUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCGAACUGCAGCAGAG----------CACCUCCU---CUCACC .((((.........(....).((((.((((((.......))))))((((.....))))............))))......)----------))).....---...... ( -22.20) >consensus AGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCGAACUGCAACAGGGUCCAACACUGCACCUCCU_CUCUCACC .((((.........(....).((((.((((((.......))))))((((.....))))............))))................)))).............. (-17.76 = -18.76 + 1.00)

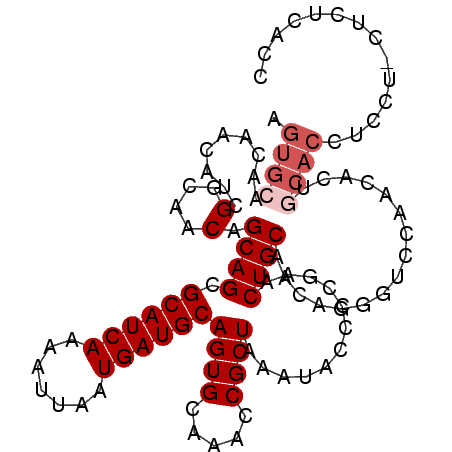
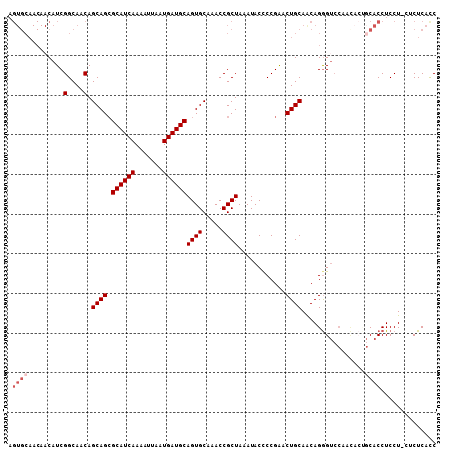
| Location | 13,190,336 – 13,190,442 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.60 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13190336 106 + 23771897 CGGGGUAUUUAGCGGUUUGCACUGCAUCAUUAAUUUUGAUGCUCUGCUGUUGCCGAUGUUGUUGCACUGGUGUGUCUGGUGUGCUGCCAAUGAAGGGAGUUGCCUU .((((((.....((((..(((..((((((.......))))))..)))....))))........((((..(.....)..))))(((.((......)).))))))))) ( -34.70) >DroSec_CAF1 41687 101 + 1 CGGAGUAUUUAGCGGUUUGCACUGCAUCAUUAAUUUUGAUGCGCUGCUGUUGCCGAUGUUGUUGCACUGGUGUC-----CGUGCUGCCAAUGAAGGGAGUUCCCUU .(((((.....((((...(((.(((((((.......))))))).)))....((((.(((....))).))))..)-----)))))).))....(((((....))))) ( -32.20) >DroSim_CAF1 40061 101 + 1 CGGAGUAUUUAGCGGUUUGCACUGCAUCAUUAAUUUUGAUGCGCUGCUGUUGCCGAUGUUGUUGCACUGGUGUC-----UGUGCUGCCACUGAAGGGAGUUCCCUU .......(((((.(((..((((.((((((.......)))))).........((((.(((....))).))))...-----.)))).))).)))))(((....))).. ( -32.60) >DroEre_CAF1 45327 84 + 1 CGGGGUAUUUAGCGGUUUGCACUGCAUCAUUAACUUUGAUGCGCUGCUGUUGCCGAUG----------------------GUGCUGCCCAUGAAGAGAGCUCCCUU .(((((.(((..((((..(((.(((((((.......))))))).)))....))))(((----------------------(......))))...))).)))))... ( -28.00) >DroYak_CAF1 41208 99 + 1 CGGGGUAUUUAGCGGUUUGCACUGCAUCAUUAACUUUGAUGCGCUGCUGUUGCCGAUGUUGUUGCACU--UGUC-----UGUGUUGCCCAUGAAGAGAGCUCCUUU .(((((.(((..((((..(((.(((((((.......))))))).)))....))))(((..((.((((.--....-----.)))).)).)))...))).)))))... ( -31.50) >consensus CGGGGUAUUUAGCGGUUUGCACUGCAUCAUUAAUUUUGAUGCGCUGCUGUUGCCGAUGUUGUUGCACUGGUGUC_____UGUGCUGCCAAUGAAGGGAGUUCCCUU .(((((((....((((..(((.(((((((.......))))))).)))....))))........(((....))).......))))).))....(((((....))))) (-22.95 = -23.34 + 0.39)

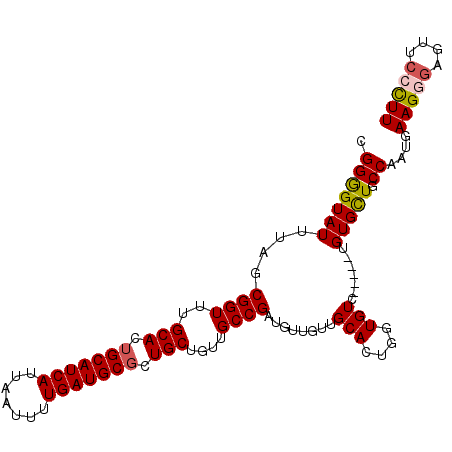
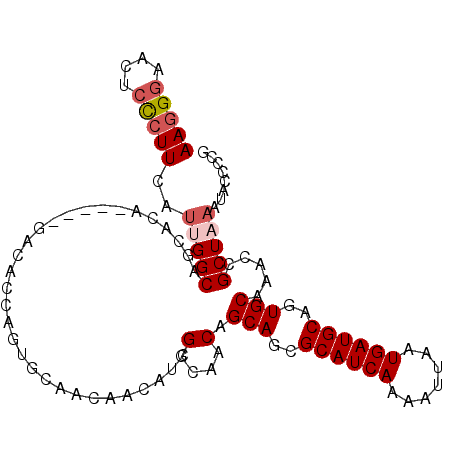
| Location | 13,190,336 – 13,190,442 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 85.60 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13190336 106 - 23771897 AAGGCAACUCCCUUCAUUGGCAGCACACCAGACACACCAGUGCAACAACAUCGGCAACAGCAGAGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCG .((....)).......(((((.((((.............)))).........(....).(((..((((((.......))))))..))).....)))))........ ( -23.52) >DroSec_CAF1 41687 101 - 1 AAGGGAACUCCCUUCAUUGGCAGCACG-----GACACCAGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACUCCG (((((....)))))..(((((.((((.-----.......)))).........(....).(((..((((((.......))))))..))).....)))))........ ( -29.40) >DroSim_CAF1 40061 101 - 1 AAGGGAACUCCCUUCAGUGGCAGCACA-----GACACCAGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACUCCG (((((....))))).(((((..((((.-----.......)))).........(....).(((..((((((.......))))))..)))...))))).......... ( -29.40) >DroEre_CAF1 45327 84 - 1 AAGGGAGCUCUCUUCAUGGGCAGCAC----------------------CAUCGGCAACAGCAGCGCAUCAAAGUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCG ..(((.((((.......))))(((..----------------------....(....).(((..((((((.......))))))..))).....)))......))). ( -26.20) >DroYak_CAF1 41208 99 - 1 AAAGGAGCUCUCUUCAUGGGCAACACA-----GACA--AGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAGUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCG ...((.((((.......))))......-----....--((((..........(....).(((..((((((.......))))))..)))....)))).......)). ( -22.10) >consensus AAGGGAACUCCCUUCAUUGGCAGCACA_____GACACCAGUGCAACAACAUCGGCAACAGCAGCGCAUCAAAAUUAAUGAUGCAGUGCAAACCGCUAAAUACCCCG (((((....)))))..(((((...............................(....).(((..((((((.......))))))..))).....)))))........ (-18.76 = -19.52 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:44 2006