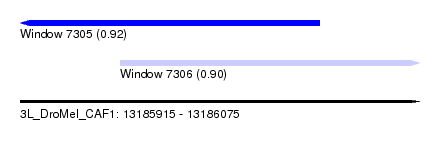
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,185,915 – 13,186,075 |
| Length | 160 |
| Max. P | 0.922930 |

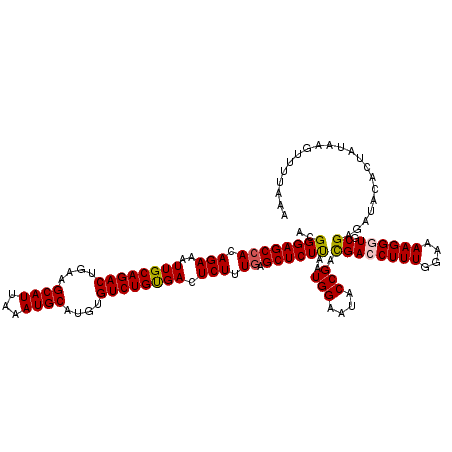
| Location | 13,185,915 – 13,186,035 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.62 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -25.86 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13185915 120 - 23771897 CGUCGGUAUUCCAUUAAGAGCUCAAAGAGUCGCAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUGCUACAUUGUGUUCCCUCCUUCU ....((....)).....((((.(((((((..((((((....((((...))))....))))))...))))(((((....((..(((.....))).)).))))).))).))))......... ( -29.70) >DroSec_CAF1 37434 117 - 1 CGUCGGUAUUCCAUUAAGAGCUCAAAGAGUCACAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUG---CAUUGUGUUCUCGCCUUCU ....((....))....(((((.(((.((((((((((....((((...............))))...))))))))))..((..(((.....))).))..---..))).)))))........ ( -27.36) >DroSim_CAF1 35675 117 - 1 CGUCGGUAUUCCAUUAAGAGCUCAAAGAGUCACAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUG---CAUUGUGUUCUCUCCUUCU ....((....))....(((((.(((.((((((((((....((((...............))))...))))))))))..((..(((.....))).))..---..))).)))))........ ( -27.36) >DroEre_CAF1 40917 117 - 1 CGUCGGUAUUCCAUUAAGAGCUCAAAGAGUCACAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUG---CAUUGUGUUCUCUCCUUCU ....((....))....(((((.(((.((((((((((....((((...............))))...))))))))))..((..(((.....))).))..---..))).)))))........ ( -27.36) >DroYak_CAF1 36598 117 - 1 CAACGGUAUUCCAUUGAGAGCUGAAAGAGUCACAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUG---CAUUGUGUUCUCUCCUUCU ....((....))...((((((.((..((((((((((....((((...............))))...))))))))))...))..(((((.((.....))---.)))))))))))....... ( -28.86) >consensus CGUCGGUAUUCCAUUAAGAGCUCAAAGAGUCACAGACACAUGCAUUUAAUGCUUCAGUCUGCAAUUUCUGUGGCUCCCGUCUGACAAUCCGUCUACUG___CAUUGUGUUCUCUCCUUCU ....((....)).....((((.(((.((((((((((....((((...............))))...))))))))))..((..(((.....))).)).......))).))))......... (-25.86 = -25.90 + 0.04)

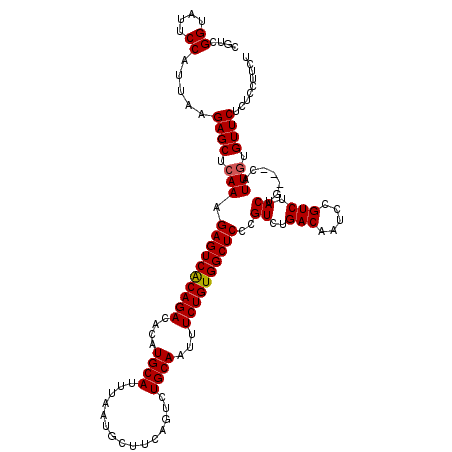
| Location | 13,185,955 – 13,186,075 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.48 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13185955 120 + 23771897 ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGCGACUCUUUGAGCUCUUAAUGGAAUACCGACGACCUUUGGAAAAGGGUGCGAGAUACACUAUAAGUUUAAAA ..((((((((.(((..((((((((....((((...))))....)))))))).))).)).))))))..(((....))).((((((((....)))))).))..................... ( -35.10) >DroSec_CAF1 37471 120 + 1 ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGUGACUCUUUGAGCUCUUAAUGGAAUACCGACGAACUUUGGAAAAGGGUGCGAGAUACACUAUAAGUUUUAAA ..((((((((.(((..(..(((((....((((...))))....)))))..).))).)).))))))..((((((.((((......)))).....((((.......))))....)))))).. ( -29.30) >DroSim_CAF1 35712 120 + 1 ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGUGACUCUUUGAGCUCUUAAUGGAAUACCGACGACCUUUGGAAAAGGGUGCGAGAUACACUAUAAGUUUUAAA ..((((((((.(((..(..(((((....((((...))))....)))))..).))).)).))))))..(((....))).((((((((....)))))).))..................... ( -32.90) >DroEre_CAF1 40954 120 + 1 ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGUGACUCUUUGAGCUCUUAAUGGAAUACCGACGAACUUUGGGAAAGGGUGCGAGAUACACUAUAAGUCUUAAA .(((((((((.(((..(..(((((....((((...))))....)))))..).))).)).)))))...(((....))).))....((((((...((((.......)))).....)))))). ( -30.60) >DroYak_CAF1 36635 120 + 1 ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGUGACUCUUUCAGCUCUCAAUGGAAUACCGUUGACCUUUGGAAAAGGGUGCGAGAUACACUACAAGUUUUCAA ..(..(((....(((((..(((((....((((...))))....)))))..)...))))..(((.((((((....))))))((((((....))))))..)))...........)))..).. ( -33.90) >consensus ACGGGAGCCACAGAAAUUGCAGACUGAAGCAUUAAAUGCAUGUGUCUGUGACUCUUUGAGCUCUUAAUGGAAUACCGACGACCUUUGGAAAAGGGUGCGAGAUACACUAUAAGUUUUAAA ..((((((((.(((..((((((((....((((...))))....)))))))).))).)).))))))..(((....))).((((((((....)))))).))..................... (-28.16 = -29.48 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:33 2006