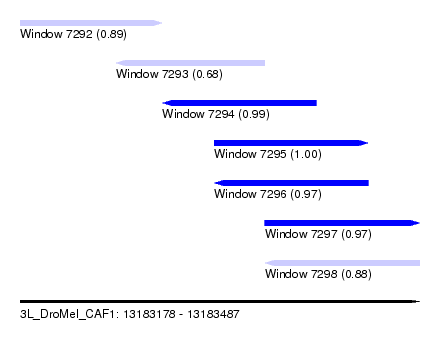
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,183,178 – 13,183,487 |
| Length | 309 |
| Max. P | 0.999934 |

| Location | 13,183,178 – 13,183,288 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -33.74 |
| Energy contribution | -34.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183178 110 + 23771897 AUAUAGAAAAUUGUUGAGGGCAGGC----GUGCGGAGGUCUAGCCACGUUUUCUGGCAGAUCCGAGCAUGAUUUAAAGGGC-GA---CGGCCCAGUCCUGUCUAUUGGCUUUGUUGUU ............((..(((((((((----((((...(((((.((((.......)))))))))...))))).......((((-..---..))))...))))))).)..))......... ( -38.30) >DroSec_CAF1 34739 111 + 1 AUAUAGAAAAUUGUUGAGGGCAGGC----GUGCGGAGAUCUAGCCACGUUUUCUGGCAGAUCCGAGCAUGAUUUAAAGGGCAGA---CGGCCCAGUCCUGUCUGUUGGCUUUGUUGUU ............((..(((((((((----((((...(((((.((((.......)))))))))...))))).......((((...---..))))...))))))).)..))......... ( -38.70) >DroSim_CAF1 32981 111 + 1 GUAUAGAAAAUUGUUGAGGGCAGGC----GUGCGGAGAUCUAGCCACGUUUUCUGGCAGAUCCGAGCAUGAUUUAAAGGGCAGA---CGGCCCAGUCCUGUCUGUUGGCUUUGUUGUU ............((..(((((((((----((((...(((((.((((.......)))))))))...))))).......((((...---..))))...))))))).)..))......... ( -38.70) >DroEre_CAF1 38245 109 + 1 AUCUAGAAAAUUGUUGAGGGCAGGCG-----GCGGAGAUCUAGCCACGUU-UCUGGCAGAUCCGAGCAUGAUUUAAAGGGCAGA---CGGCCCAGUCCAGUCUGUUGGCUUUGUUGUU .................((((..((.-----.((((..(((.((((....-..))))))))))).))..........((((...---..)))).))))((((....))))........ ( -32.00) >DroYak_CAF1 33946 117 + 1 -UAUAGAAAAUUGUUGAGGGCAGGCAGGCGUGCGGAGAUCUAGCCACGUUUUCUGGCAGAUCCGAGCAUGAUUUAAAGGGCAGACGACGGCCCAGUCCUGUCUGUUGGCUUUGUUGUU -..............(..(((((((((((((((...(((((.((((.......)))))))))...))))).......((((........))))...))))))))))..)......... ( -41.50) >consensus AUAUAGAAAAUUGUUGAGGGCAGGC____GUGCGGAGAUCUAGCCACGUUUUCUGGCAGAUCCGAGCAUGAUUUAAAGGGCAGA___CGGCCCAGUCCUGUCUGUUGGCUUUGUUGUU ............((..((((((((.....((((...(((((.((((.......)))))))))...))))........((((........))))...))))))).)..))......... (-33.74 = -34.18 + 0.44)

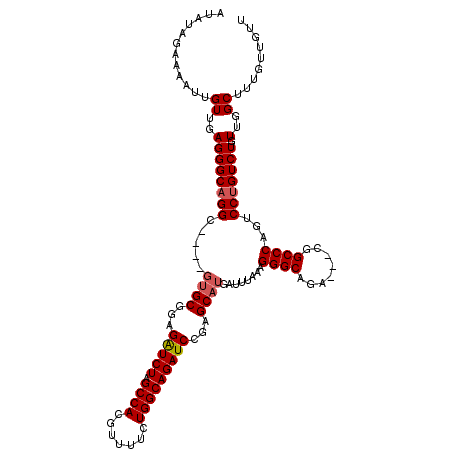
| Location | 13,183,252 – 13,183,367 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183252 115 - 23771897 CAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGAAUCAACAAAGGCUUACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAAUAGACAGGACUGGGCCG---UC-GCC ....(((((..(((...-.....))).)))))....((((.........(((((.........(((((......)))))........)))))........((......)).---))-)). ( -21.33) >DroSec_CAF1 34813 116 - 1 CAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAACAGACAGGACUGGGCCG---UCUGCC .....(((((.......-......(((((..(((.((.(((((......))))).)).)))....)))))...................(((..(((......))))))..---))))). ( -27.80) >DroSim_CAF1 33055 116 - 1 CAAAUGCAUAGACUACA-ACUACAGUGCUGCAGAAUUCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAACAGACAGGACUGGGCCG---UCUGCC ........(((((....-......(((((..(((....(((((......)))))....)))....)))))...................(((..(((......)))))).)---)))).. ( -22.20) >DroEre_CAF1 38317 117 - 1 CAAAUGCAGAGGCUACACGCUACAGUGUUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAACAGACUGGACUGGGCCG---UCUGCC .....(((((((((....(((....(((((.(((.((.(((((......))))).)).)))..(((((......))))))))))....)))...(((......))))))).---))))). ( -34.00) >DroYak_CAF1 34023 119 - 1 CAAAUGAGGAGACUGCA-ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAACAGACAGGACUGGGCCGUCGUCUGCC ......((..(((.((.-....((((.(((((((.((.(((((......))))).)).)))..(((((......))))).................).))).)))).)).)))..))... ( -29.50) >consensus CAAAUGCAGAGACUACA_ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUCAACAACAAAGCCAACAGACAGGACUGGGCCG___UCUGCC .....(((((..............(((((..(((.((.(((((......))))).)).)))....)))))...................(((..(((......)))))).....))))). (-20.86 = -21.26 + 0.40)



| Location | 13,183,288 – 13,183,407 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183288 119 - 23771897 CUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGAAUCAACAAAGGCUUACAAUCUAUAAAGUACAAUCACUUUC ..(((...(((....((((....(((((....))))).))))..)))..))).....-......(((((..(((.((..(..(......)..)..)).)))....))))).......... ( -22.90) >DroSec_CAF1 34850 119 - 1 CUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUC ..(((...(((....((((....(((((....))))).))))..)))..))).....-......(((((..(((.((.(((((......))))).)).)))....))))).......... ( -30.10) >DroSim_CAF1 33092 119 - 1 CUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAUAGACUACA-ACUACAGUGCUGCAGAAUUCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUC ..((((..(((....((((....(((((....))))).))))..))).)))).....-......(((((..(((....(((((......)))))....)))....))))).......... ( -26.00) >DroEre_CAF1 38354 120 - 1 CUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGGCUACACGCUACAGUGUUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUC ...((...(((....((((....(((((....))))).))))..)))..))((.((((......)))).))(((.((.(((((......))))).)).)))..(((((......))))). ( -31.10) >DroYak_CAF1 34063 119 - 1 CUUCUAAGGCAAACAUGCCCGACACGUUGCACAACGUUGGCAAAUGAGGAGACUGCA-ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUC .......((((....)))).((...(((((....((((....))))((....)))))-))....(((((..(((.((.(((((......))))).)).)))....)))))..))...... ( -31.70) >consensus CUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA_ACUACAGUGCUGCAGAAUGCGAGUCAACAAAGGCUCACAAUCUAUAAAGUACAAUCACUUUC ..(((...(((....((((....(((((....))))).))))..)))..)))............(((((..(((.((.(((((......))))).)).)))....))))).......... (-25.14 = -25.26 + 0.12)

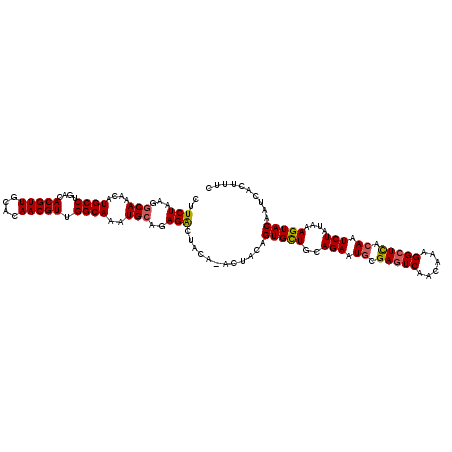
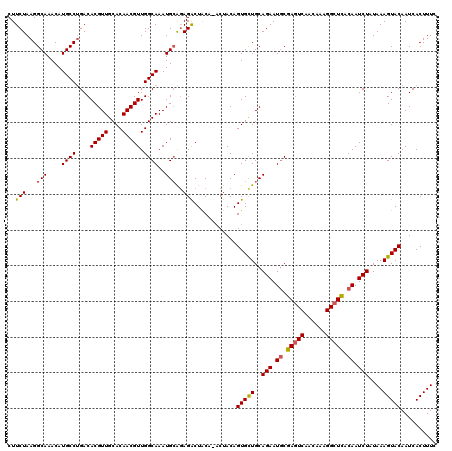
| Location | 13,183,328 – 13,183,447 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -48.92 |
| Consensus MFE | -44.04 |
| Energy contribution | -43.96 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183328 119 + 23771897 UCGCAUUCUGCAGCACUGUAGU-UGUAGUCUCUGCAUUUGCCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAUGU ..((((((((((((((.((((.-(((((...))))).)))).......))))..((((.((((((....)))))(((((((((((.....)))))))))))).))))...)))))))))) ( -51.90) >DroSec_CAF1 34890 119 + 1 UCGCAUUCUGCAGCACUGUAGU-UGUAGUCUCUGCAUUUGCCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCAUUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGU .......(((((((......))-)))))..(((((((((((((....)).))..((((.((((((....)))))(((((((((((.....)))))))))))).))))))))))))).... ( -48.40) >DroSim_CAF1 33132 119 + 1 UCGAAUUCUGCAGCACUGUAGU-UGUAGUCUAUGCAUUUGCCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGU .....(((((((((((....((-(((((.........))).))))...))))..((((.((((((....)))))(((((((((((.....)))))))))))).))))...)))))))... ( -46.50) >DroEre_CAF1 38394 120 + 1 UCGCAUUCUGCAACACUGUAGCGUGUAGCCUCUGCAUUUGCCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCGGAAAGU ((((((((((((.((.(((.((((((((...))))))..))..))).)))))).((((.((((((....)))))(((((((((((.....)))))))))))).))))))))))))..... ( -49.20) >DroYak_CAF1 34103 119 + 1 UCGCAUUCUGCAGCACUGUAGU-UGCAGUCUCCUCAUUUGCCAACGUUGUGCAACGUGUCGGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAGCUU ..((..((((((((((....((-(((((.........))).))))...))))..((((.((((((....)))))(((((((((((.....)))))))))))).))))...)))))))).. ( -48.60) >consensus UCGCAUUCUGCAGCACUGUAGU_UGUAGUCUCUGCAUUUGCCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGU .....(((((((...((((....((((.....))))...(((.(((((....)))))...(((((....))))).((((((((((.....))))))))))))).))))..)))))))... (-44.04 = -43.96 + -0.08)

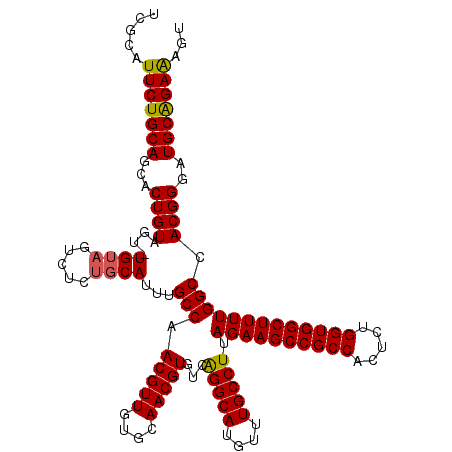

| Location | 13,183,328 – 13,183,447 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.72 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183328 119 - 23771897 ACAUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGA .(((((((((...(((((((....(((((....)))))))).....(((((....)))))..))))..((((...((((((....))((...)).))-))....)))))))))))))... ( -41.10) >DroSec_CAF1 34890 119 - 1 ACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAAUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA-ACUACAGUGCUGCAGAAUGCGA ......((((...((((.....(((((.((.....)).)))))...(((((....)))))..)))).))))........(((..(((((..(((...-.....))).)))))...))).. ( -36.20) >DroSim_CAF1 33132 119 - 1 ACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAUAGACUACA-ACUACAGUGCUGCAGAAUUCGA ...(((((((...(((((((....(((((....)))))))).....(((((....)))))..))))..((((...((((................))-))....)))))))))))..... ( -37.29) >DroEre_CAF1 38394 120 - 1 ACUUUCCGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGGCUACACGCUACAGUGUUGCAGAAUGCGA ......(((((.(.((((((..(((((.((.....)).))))).....(((....((((....(((((....))))).))))..)))...))))))((((....))))....).))))). ( -40.20) >DroYak_CAF1 34103 119 - 1 AAGCUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCCGACACGUUGCACAACGUUGGCAAAUGAGGAGACUGCA-ACUACAGUGCUGCAGAAUGCGA ..((((((((((((((.((((.(((((.((.....)).)))))....((((....))))....(((((....)))))))))..))).))).((((..-....))))..))))))..)).. ( -40.00) >consensus ACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGGCAAAUGCAGAGACUACA_ACUACAGUGCUGCAGAAUGCGA ...(((((((.((.((((((..(((((.((.....)).)))))....))).....((((....(((((....))))).))))..................))).).).)))))))..... (-32.32 = -32.72 + 0.40)

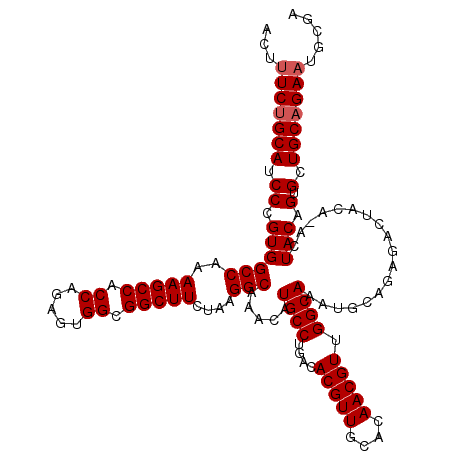

| Location | 13,183,367 – 13,183,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -48.62 |
| Consensus MFE | -46.26 |
| Energy contribution | -45.94 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183367 120 + 23771897 CCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAUGUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGGCAGC ((((((((.((((.((((.((((((....)))))(((((((((((.....)))))))))))).))))...)))).))))).(((((((((((....)).)))...))).))).))).... ( -48.90) >DroSec_CAF1 34929 120 + 1 CCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCAUUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGGCAGC .....((..((((.......(((((....)))))....(((((((.((((((.(((.((((.((((............)))))))).))))))))))))))))......))))..))... ( -47.70) >DroSim_CAF1 33171 120 + 1 CCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGGCAGC .....((..((((..((((((((((....)))))(((((((((((.....)))))))))))(((((............))))).)))))(((((.......)))))...))))..))... ( -47.30) >DroEre_CAF1 38434 120 + 1 CCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCGGAAAGUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGGCAGC .....((..(((..(((((((((((....)))))(((((((((((.....))))))))))).......))))))....(((((....(((((....)).)))...))))))))..))... ( -48.70) >DroYak_CAF1 34142 120 + 1 CCAACGUUGUGCAACGUGUCGGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAGCUUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGACAGC .....(((((....((((.((((((....)))))(((((((((((.....)))))))))))).))))..(((((..(((..((....((((....)))).))..)))..))))).))))) ( -50.50) >consensus CCAACGUUGUGCAACGUGUCAGGCAUGUUUGCCUUAGAAGCCGCCACUCUGGUGGCUUUUGGCCACGGGAUGCAGAAAGUGGCAGAUGCCUCGGGAGGUGGUUGAGUCAUGCAUGGCAGC .....((..((((.((((.((((((....)))))(((((((((((.....)))))))))))).))))............((((....(((((....)).)))...))))))))..))... (-46.26 = -45.94 + -0.32)

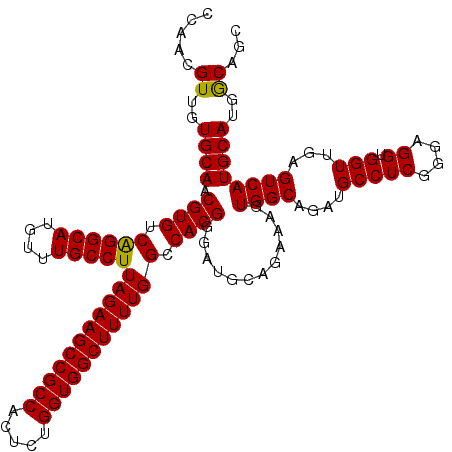
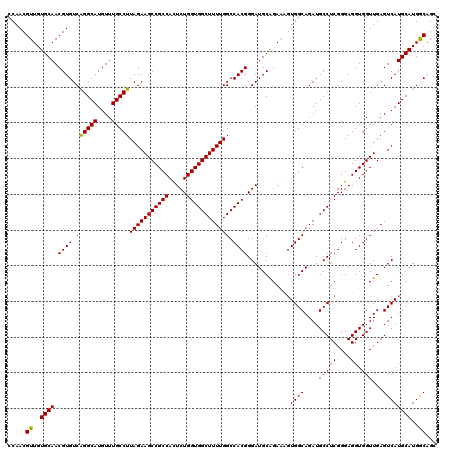
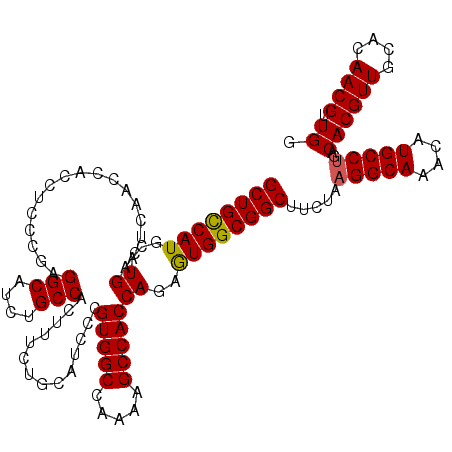
| Location | 13,183,367 – 13,183,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -36.40 |
| Energy contribution | -36.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13183367 120 - 23771897 GCUGCCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCACAUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGG (((((((((((.((.((...........))(((....)))....))))).((..(((((.....)))))..)))))))))).....(((((....)))))..((((((....)))).)). ( -37.70) >DroSec_CAF1 34929 120 - 1 GCUGCCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAAUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGG (((((((((((.((.((...........))(((....)))....))))).((..(((((.....)))))..)))))))))).....(((((....)))))..((((((....)))).)). ( -37.40) >DroSim_CAF1 33171 120 - 1 GCUGCCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGG (((((((((((.((.((...........))(((....)))....))))).((..(((((.....)))))..)))))))))).....(((((....)))))..((((((....)))).)). ( -37.70) >DroEre_CAF1 38434 120 - 1 GCUGCCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCACUUUCCGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGG ((((((((...((.................(((....)))..............(((((.....)))))))..)))))))).....(((((....)))))..((((((....)))).)). ( -37.50) >DroYak_CAF1 34142 120 - 1 GCUGUCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCAAGCUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCCGACACGUUGCACAACGUUGG ...((((....))))....(((.....((.(((((.((((((((((..(.((..(((((.....)))))..)))..).)))))....))))...)))))))..(((((....)))))))) ( -37.40) >consensus GCUGCCAUGCAUGACUCAACCACCUCCCGAGGCAUCUGCCACUUUCUGCAUCCCGUGGCCAAAAGCCACCAGAGUGGCGGCUUCUAAGGCAAACAUGCCUGACACGUUGCACAACGUUGG ((((((((...((.................(((....)))..............(((((.....)))))))..)))))))).....(((((....)))))..((((((....)))).)). (-36.40 = -36.28 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:25 2006