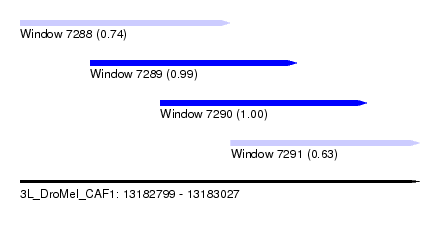
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,182,799 – 13,183,027 |
| Length | 228 |
| Max. P | 0.999924 |

| Location | 13,182,799 – 13,182,919 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -34.10 |
| Energy contribution | -33.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13182799 120 + 23771897 UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCACCUUCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAA ...(((...((........((((((........(((.((.......)).)))...........))))))........))...((((((((((((....)).))))))))))....))).. ( -34.70) >DroSec_CAF1 34363 120 + 1 UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCACCUUCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCGUGCCACUUUAGUGGGCGUGCUGCAAAACGUAA ...(((((((((((.....)).)))....................(((((.(((.(((....(((.(((....))).)))))).))).))))).......)))))).............. ( -33.90) >DroSim_CAF1 32599 120 + 1 UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCACCUUCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAA ...(((...((........((((((........(((.((.......)).)))...........))))))........))...((((((((((((....)).))))))))))....))).. ( -34.70) >DroEre_CAF1 37880 119 + 1 UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCGCCUUCCUUUGGCGAGCCUCCUCUGCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAA-AGGUAA ..((((.((((((((......))).........((((((.......))))))........))))).))))......(((...((((((((((((....)).))))))))))..-.))).. ( -43.90) >DroYak_CAF1 33581 120 + 1 UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCACCUUCCUUUGGAGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCCACAAAACGUAA ...((((...))))....(((((((((..............((...((((((....)))))).))..............)))))((((((((((....)).))))))))......)))). ( -36.19) >consensus UCAACGCCCGGCGUCAUCUGCGGCCUUUAUCAAUUCACCUUCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAA ...((((((((((.....(((.(((((.......((.((.......)).))(((.........))).............))))).)))))))((....)))))))).............. (-34.10 = -33.94 + -0.16)

| Location | 13,182,839 – 13,182,957 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13182839 118 + 23771897 UCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAGAAGAAAAUACUAUUAAUAAGAAAAAAUAAGAA--AAA ..((((.(((((((.........))))...............((((((((((((....)).))))))))))....))).))))................................--... ( -30.70) >DroSec_CAF1 34403 117 + 1 UCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCGUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAGAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAA ..((((.(((((((.........))))...............((((((((((((....)).))))))))))....))).))))..................-.............--... ( -27.30) >DroSim_CAF1 32639 117 + 1 UCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAUAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAG ..(((..(((((((.........))))...............((((((((((((....)).))))))))))....))).....)))...............-.............--... ( -28.20) >DroEre_CAF1 37920 115 + 1 UCCUUUGGCGAGCCUCCUCUGCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAA-AGGUAAAA---GAAAAUACUAUUAAUA-CAAAAAAUAAGAAACAAA ..(((((((.(((.......))).))).........(((...((((((((((((....)).))))))))))..-.))).)))---)...............-.................. ( -32.10) >DroYak_CAF1 33621 114 + 1 UCCUUUGGAGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCCACAAAACGUAAAA---GAAAAUACUAUUAAUG-CAAAAAAUGAGAA--AAA ..((((((..((((.........))))(....)....)))))).((((((((((....)).)))))))).............---................-.............--... ( -26.60) >consensus UCCUUUGGCGAGCCUCCUCUUCUGGCUGUUUACAUAACCAAGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAA_AAGAAAAUACUAUUAAUA_CAAAAAAUAAGAA__AAA ...(((((..((((.........))))(....)....)))))((((((((((((....)).))))))))))................................................. (-25.66 = -25.54 + -0.12)


| Location | 13,182,879 – 13,182,997 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -23.20 |
| Energy contribution | -22.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13182879 118 + 23771897 AGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAGAAGAAAAUACUAUUAAUAAGAAAAAAUAAGAA--AAAAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGA ..((((((((((((....)).))))))))))............................................--....(((((.(((((......)))))..))))).......... ( -25.80) >DroSec_CAF1 34443 117 + 1 AGGCGGCGUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAGAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAAAGUAUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGA ...((((((((..((((.....((((........))))....)))).........((((((-((((.........--............)))))))))).......))))).)))..... ( -23.90) >DroSim_CAF1 32679 117 + 1 AGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAAUAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAGAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGA ..((((((((((((....)).)))))))))).....................(((((.(((-(............--....)))).....((((((..........))))))..))))). ( -26.59) >DroEre_CAF1 37960 115 + 1 AGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAA-AGGUAAAA---GAAAAUACUAUUAAUA-CAAAAAAUAAGAAACAAAAGUGCAAAAUUUGUUCUAAAAUUAAUGCACGACUGGUAGA ..((((((((((((....)).))))))))))..-.((((...---.....)))).......-..............((...(((((.(((((......)))))..)))))...))..... ( -29.00) >DroYak_CAF1 33661 114 + 1 AGGCGGCAUGCCACUUUAGUGGGCGUGCCACAAAACGUAAAA---GAAAAUACUAUUAAUG-CAAAAAAUGAGAA--AAAAGUGAAAAAUUUGUGUUAAAAUUAAUGCACGACUAGUAGA ..(.((((((((((....)).)))))))).)...........---.....(((((.(..((-((...(((...((--..((((.....))))...))...)))..))))..).))))).. ( -24.10) >consensus AGGCGGCAUGCCACUUUAGUGGGCGUGCUGCAAAACGUAAAA_AAGAAAAUACUAUUAAUA_CAAAAAAUAAGAA__AAAAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGA ..((((((((((((....)).)))))))))).....................(((((......................((((.....))))((((..........))))....))))). (-23.20 = -22.80 + -0.40)

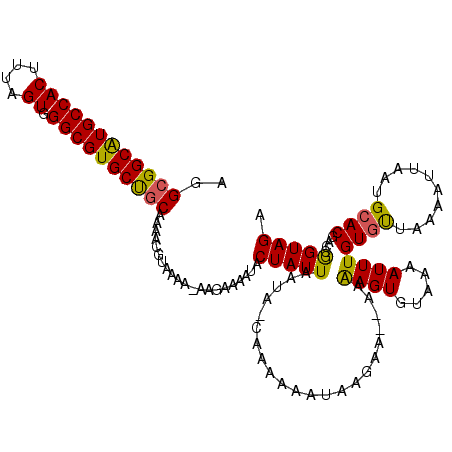
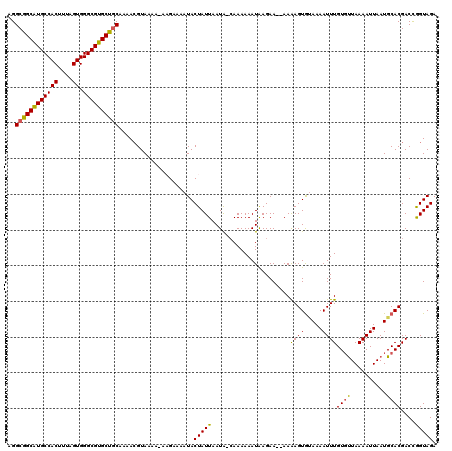
| Location | 13,182,919 – 13,183,027 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -11.84 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13182919 108 + 23771897 AAGAAGAAAAUACUAUUAAUAAGAAAAAAUAAGAA--AAAAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGAGAAGGCUGCGCCUUCC----------CUACGCUAAGUAAA ..........((((.....................--....(((((.(((((......)))))..))))).....((((.((((((...)))))).----------))))....)))).. ( -21.10) >DroSec_CAF1 34483 107 + 1 AAGAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAAAGUAUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGAGAAGGCUGCGCCUUCC----------CUACGCUAAGUAAA ..........((((.((((((-((((.........--............)))))))))).......((.(....)((((.((((((...)))))).----------))))))..)))).. ( -20.90) >DroSim_CAF1 32719 107 + 1 AAUAAGAAAAUACUAUUAAUA-CAAAAAAUAAGAA--AAGAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGAGGAGGCUGCGCCUUCC----------CUACGCUAAGUAAA ..........((((.......-.............--....(((((.(((((......)))))..))))).....((((.((((((...)))))).----------))))....)))).. ( -20.60) >DroEre_CAF1 37999 90 + 1 AA---GAAAAUACUAUUAAUA-CAAAAAAUAAGAAACAAAAGUGCAAAAUUUGUUCUAAAAUUAAUGCACGACUGGUAGA--------------------------CUGCGCUAAGUAAA ..---.....((((.......-...................(((((.(((((......)))))..)))))...((((...--------------------------....)))))))).. ( -10.10) >DroYak_CAF1 33701 114 + 1 AA---GAAAAUACUAUUAAUG-CAAAAAAUGAGAA--AAAAGUGAAAAAUUUGUGUUAAAAUUAAUGCACGACUAGUAGAGAAGGCUGUGCCUUCCCUUCCCCUCCCCACGCUAAGUAAA ..---.....((((.......-((.....))....--...((((......((((((..........))))))...(.((.((((((...)))))).)).).........)))).)))).. ( -15.80) >consensus AA_AAGAAAAUACUAUUAAUA_CAAAAAAUAAGAA__AAAAGUGUAAAAUUUGUGUUAAAAUUAAUGCACGACCGGUAGAGAAGGCUGCGCCUUCC__________CUACGCUAAGUAAA ........................................((((((....((((((..........))))))........((((((...))))))............))))))....... (-11.84 = -12.42 + 0.58)

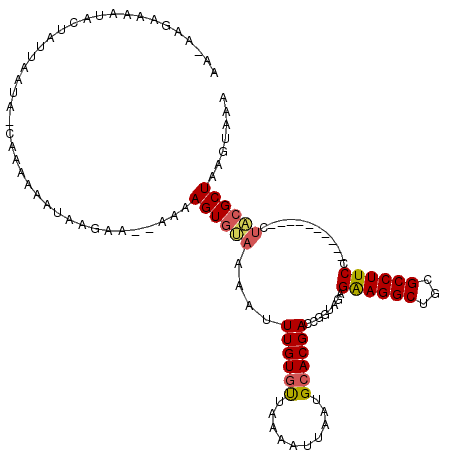
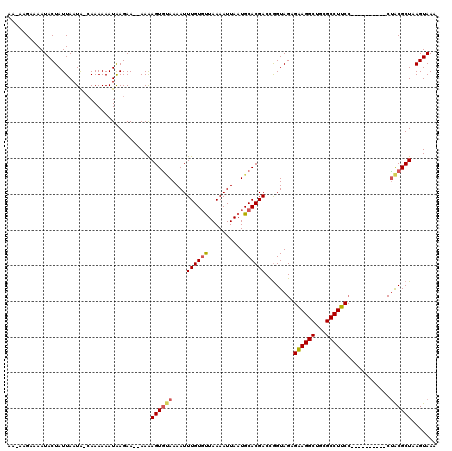
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:19 2006