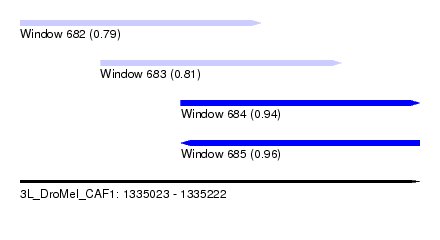
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,335,023 – 1,335,222 |
| Length | 199 |
| Max. P | 0.963846 |

| Location | 1,335,023 – 1,335,143 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -31.84 |
| Energy contribution | -33.00 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1335023 120 + 23771897 GGAGAGCAGAAAUUUAAGCCUUUGUUUCCCUUUGCAGGAGGUGUUCGCGGAGUGUUCGACGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUAC ....((((((...................((((((.((((..((((((.((((.((((....)))).)))).)).))))..)))))))))).(((((((.....))))))).)))))).. ( -43.60) >DroSec_CAF1 7264 120 + 1 GGAGAGCUGAAAUUAAAGUCUUUGUUUGCCUUUGCAGGAGGCGUUCGCGGAGUGUUUGACGUUGAGGACUUGGUGGAGCUGCUCCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUAC ((((((((..(....(((((((((..((((((.....))))))..)(((..(....)..))).))))))))..)..)))).))))(((.(..(((((((.....)))))))..))))... ( -39.50) >DroSim_CAF1 7321 120 + 1 GGAGAGCUGAAAUUAAAGUCUUUGUUUGCCUUUGCAGGAGGCGUUCGCGGAGUGUUCGACGUUGAGGACUUGGUGGAGCUGCUGCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUAC (((((.((........)))))))(((..(((((((((..(((.(((((.((((.((((....)))).)))).)))))))).))))).))))..)))(..(((((.....)))))..)... ( -43.20) >DroEre_CAF1 7321 119 + 1 GGAGAGCUUCAAUUUAAGGC-UUGUGUCCCUUUGCAGGAGGCGUUCGCGGAGUGUUUGAUGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUAC (((((((((.......))))-))...)))((((((.((((((.(((((.((((.(((......))).)))).)))))))..)))))))))).....(..(((((.....)))))..)... ( -42.40) >DroYak_CAF1 7552 120 + 1 AGAGAGUUUCGAUUUAUGGCGUUGUUUCCCUUUGCAGGUGGAGUUCGCGGAGUUUUUGAUGUUGAGGACUUGGUUGAGCUGCUCCGAAAGGAGAAUGUGGACGAUAUAUUCGUCUGCUAC ................((((...(((..(((((...((.(.(((((((.((((((((......)))))))).).)))))).).)).)))))..)))..((((((.....)))))))))). ( -37.80) >consensus GGAGAGCUGAAAUUUAAGGCUUUGUUUCCCUUUGCAGGAGGCGUUCGCGGAGUGUUUGACGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUAC ....(((..........((((((((........))).))))).(((((.((((.(((......))).)))).)))))))).((((....))))...(..(((((.....)))))..)... (-31.84 = -33.00 + 1.16)

| Location | 1,335,063 – 1,335,183 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -37.26 |
| Energy contribution | -38.30 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1335063 120 + 23771897 GUGUUCGCGGAGUGUUCGACGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUACGUUGACCACCUGGUGGUUU ...(((((.((((.((((....)))).)))).)))))....((((....))))...(..(((((.....)))))..).(((((((((.....)))..)))))).((((((...)))))). ( -43.50) >DroSec_CAF1 7304 120 + 1 GCGUUCGCGGAGUGUUUGACGUUGAGGACUUGGUGGAGCUGCUCCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUAGUUU ((.(((((.((((.(((......))).)))).)))))))..((((....))))...(..(((((.....)))))..).(((((((((.....)))..))))))..(((....)))..... ( -38.30) >DroSim_CAF1 7361 120 + 1 GCGUUCGCGGAGUGUUCGACGUUGAGGACUUGGUGGAGCUGCUGCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUAGUUU ((.(((((.((((.((((....)))).)))).))))))).(((((...(((.(((((((.....)))))))(((....(((((((((.....)))..)))))).)))..))).))))).. ( -39.50) >DroEre_CAF1 7360 120 + 1 GCGUUCGCGGAGUGUUUGAUGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUGGACCACCUGGUGGUCU ((.(((((.((((.(((......))).)))).)))))))..((((....))))...(..(((((.....)))))..)((((((((((.....)))..)))))))((((((...)))))). ( -46.30) >DroYak_CAF1 7592 120 + 1 GAGUUCGCGGAGUUUUUGAUGUUGAGGACUUGGUUGAGCUGCUCCGAAAGGAGAAUGUGGACGAUAUAUUCGUCUGCUACGUGCCGGAAAACCUGAAGUAUGUGGACCACCUGGUGGUCU .(((((((.((((((((......)))))))).).)))))).((((....))))...(..(((((.....)))))..)((((((((((.....)))..)))))))((((((...)))))). ( -47.30) >consensus GCGUUCGCGGAGUGUUUGACGUUGAGGACUUGGUGGAGCUGCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUGGUUU ((.(((((.((((.(((......))).)))).)))))))..((((....))))...(..(((((.....)))))..).(((((((((.....)))..)))))).((((((...)))))). (-37.26 = -38.30 + 1.04)

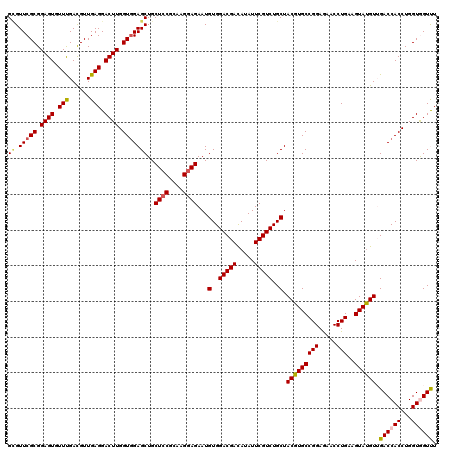
| Location | 1,335,103 – 1,335,222 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.97 |
| Mean single sequence MFE | -48.84 |
| Consensus MFE | -38.74 |
| Energy contribution | -39.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1335103 119 + 23771897 GCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUACGUUGACCACCUGGUGGUUUGCAGUGGUCGCAGUUACCG-ACACAUGCUGUCCACCGCGG ...((((.(((.(((((((.....)))))))(((....(((((((((.....)))..)))))).)))..)))(((((...(((...((((.......))-))...)))...))))))))) ( -45.90) >DroSec_CAF1 7344 119 + 1 GCUCCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUAGUUUGCAGUGGUCGCAGCUACCG-GCACAUGCUGUCCACGGCGG .((((....))))...(..(((((.....)))))..)...(((((((((...........(((.((((((.((........))))))))))).)).)))-)))).(((((....))))). ( -48.30) >DroSim_CAF1 7401 120 + 1 GCUGCGUAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUAGUUUGCAGUGGUCGCAGCUACCGGGCACAUGCUGUCCACGGCGG ...(((((.((((((((((.....))))))).)))..)))))((((..((......(((((((.(....((((((((((.((.......)))))))))))))))))))).))..)))).. ( -47.00) >DroEre_CAF1 7400 119 + 1 GCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUGGACCACCUGGUGGUCUGCAGUGGACGCAGCUACCG-ACACAUGCUGACCACGGCGG .((((....))))...(..(((((.....)))))..)....((((...............((..((((((...))))))..))((((...((((.....-......)))).)))))))). ( -48.80) >DroYak_CAF1 7632 119 + 1 GCUCCGAAAGGAGAAUGUGGACGAUAUAUUCGUCUGCUACGUGCCGGAAAACCUGAAGUAUGUGGACCACCUGGUGGUCUGCAGUGGACGCAGCUACCG-GCACAUGCUGACCACGGCGG .((((....))))...(..(((((.....)))))..)...(((((((.....(((..((.((..((((((...))))))..))....)).)))...)))-)))).(((((....))))). ( -54.20) >consensus GCUCCGCAAGGAGAAUGUGGACGACAUAUUCGUCUGCUACGUGCCGGAGAACCUGAAGUAUGUUGACCACCUGGUGGUUUGCAGUGGUCGCAGCUACCG_GCACAUGCUGUCCACGGCGG .((((....))))...(..(((((.....)))))..).(((((((((.....)))..)))))).((((((...))))))(((.((((...((((............)))).)))).))). (-38.74 = -39.58 + 0.84)

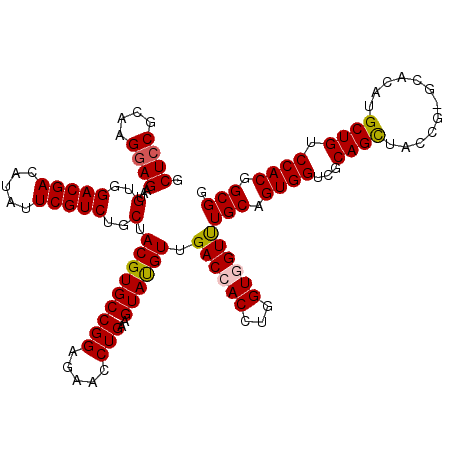
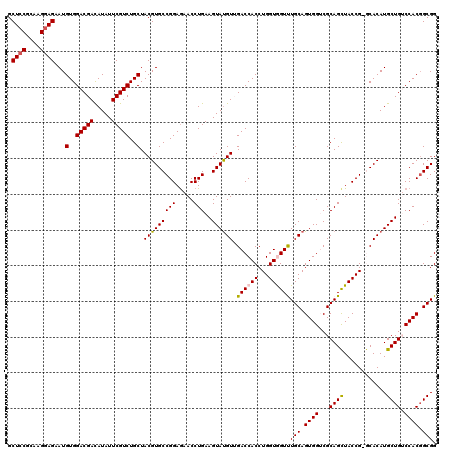
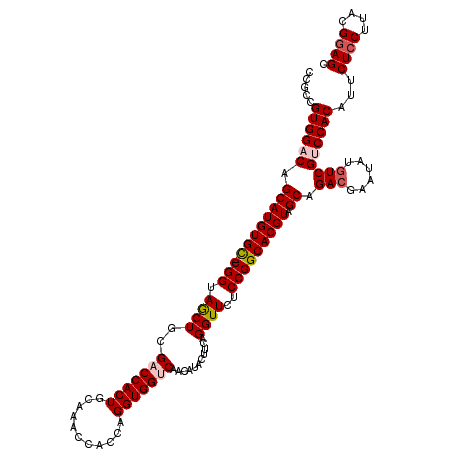
| Location | 1,335,103 – 1,335,222 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.97 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -36.02 |
| Energy contribution | -36.82 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1335103 119 - 23771897 CCGCGGUGGACAGCAUGUGU-CGGUAACUGCGACCACUGCAAACCACCAGGUGGUCAACGUACUUCAGGUUCUCCGGCACGUAGCAGACGAAUAUGUCGUCCACAUUCUCCUUGCGGAGC (((((((((((.((((((((-(((.((((..(((((((...........)))))))...(.....).))))..))))))))).)).(((......)))))))))........)))))... ( -46.00) >DroSec_CAF1 7344 119 - 1 CCGCCGUGGACAGCAUGUGC-CGGUAGCUGCGACCACUGCAAACUACCAGGUGGUCAACAUACUUCAGGUUCUCCGGCACGUAGCAGACGAAUAUGUCGUCCACAUUCUCCUUACGGAGC .....((((((.((((((((-(((.((..(((((((((...........)))))))......(....))).))))))))))).)).(((......)))))))))...((((....)))). ( -47.60) >DroSim_CAF1 7401 120 - 1 CCGCCGUGGACAGCAUGUGCCCGGUAGCUGCGACCACUGCAAACUACCAGGUGGUCAACAUACUUCAGGUUCUCCGGCACGUAGCAGACGAAUAUGUCGUCCACAUUCUCCUUACGCAGC .....((((((.(((((((((.((((.....(((((((...........)))))))....))))...((....))))))))).)).(((......)))))))))................ ( -40.30) >DroEre_CAF1 7400 119 - 1 CCGCCGUGGUCAGCAUGUGU-CGGUAGCUGCGUCCACUGCAGACCACCAGGUGGUCCACAUACUUCAGGUUCUCCGGCACGUAGCAGACGAAUAUGUCGUCCACAUUCUCCUUGCGGAGC .....((((.(.((((((.(-((...(((((((((..((..((((((...))))))..)).......((....)))).)))))))...))))))))).).))))...((((....)))). ( -44.20) >DroYak_CAF1 7632 119 - 1 CCGCCGUGGUCAGCAUGUGC-CGGUAGCUGCGUCCACUGCAGACCACCAGGUGGUCCACAUACUUCAGGUUUUCCGGCACGUAGCAGACGAAUAUAUCGUCCACAUUCUCCUUUCGGAGC .....(((....((((((((-(((.(((((((.....))).((((((...))))))...........))))..))))))))).)).(((((.....))))))))...((((....)))). ( -43.40) >consensus CCGCCGUGGACAGCAUGUGC_CGGUAGCUGCGACCACUGCAAACCACCAGGUGGUCAACAUACUUCAGGUUCUCCGGCACGUAGCAGACGAAUAUGUCGUCCACAUUCUCCUUACGGAGC .....((((((.((((((((.(((.((((..(((((((...........)))))))...........))))..))))))))).)).(((......)))))))))...((((....)))). (-36.02 = -36.82 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:35 2006