
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,177,870 – 13,177,979 |
| Length | 109 |
| Max. P | 0.906346 |

| Location | 13,177,870 – 13,177,979 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
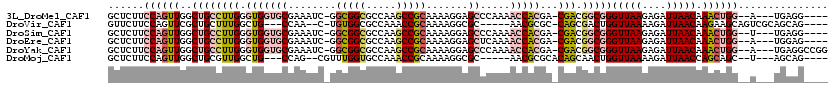
>3L_DroMel_CAF1 13177870 109 - 23771897 GCUCUUCCAGUUGGCUGCCUUGGGUGGUGCGAAAUC-GGCGGCGCCAAGCCGCAAAAGGAGCCCAAAACCACGA-CGACGGCGGGUUAAGAGAUUAACAAACUGG--A---UGAGG---- .(((.(((((((..((((((((.(((((((......-.(((((.....))))).......)).....)))))..-))).)))))(((((....))))).))))))--)---.))).---- ( -45.34) >DroVir_CAF1 61123 104 - 1 GUUCUUCCAGUUCGCUGCUUUGGCUG---CCAA--C-UGUGGCGCCAAACCGCAAAAGGCGC-----AACGCGC-CAGCGACUGGUUAAAAGAUUAACAAGAAGCAGUCGCAGCAG---- .(((((.((((.((((((((((((.(---(((.--.-..))))))))))..))....(((((-----...))))-)))))))))(((((....))))))))))((....)).....---- ( -38.60) >DroSim_CAF1 27621 109 - 1 GCUCUUCCAGUUGGCUGCCUUGGGUGGUGCGAAAUC-GGCGGCGCCAAGCCGCAAAAGGAGCCCAAAACCACGA-CGACGGCGGGUUAAGAGAUUAACAAACUGG--U---UGAGG---- .(((..((((((..((((((((.(((((((......-.(((((.....))))).......)).....)))))..-))).)))))(((((....))))).))))))--.---.))).---- ( -43.44) >DroEre_CAF1 33224 109 - 1 GCUCUUCCAGUUGGCUGCCUUGGGUGGUGCGAAAUC-GGCGGCGCCAAGCCGCAAAAGGAGCUCAAAACCACGA-CGACGGCGGGUUAAGAGAUUAACAAACUGG--A---UGGAG---- .(((((((((((..((((((((.(((((..((..((-.(((((.....))))).....))..))...)))))..-))).)))))(((((....))))).))))))--)---.))))---- ( -44.40) >DroYak_CAF1 28385 113 - 1 GCUCUUCCAGUUGGCUGCCUUGGGUGGUGCGAAAUC-GGCGGCGCCAAGCCGCAAAAGGAGCCCAAAACCACGA-CGACGGCGGGUUAAGAGAUUAACAAACUGG--A---UGAGGCCGG .(((.(((((((..((((((((.(((((((......-.(((((.....))))).......)).....)))))..-))).)))))(((((....))))).))))))--)---.)))..... ( -45.34) >DroMoj_CAF1 77995 101 - 1 GCUCUUCCAGUUGGCUGCGUUGGCUG---CCAG--CGUUUGGUGCCAAACCGCAAAAGGCGC-----AACGCGCACAGCAACUGGUUAAAAGAUUAACCAGCAGC--U---AGCAG---- .........(((((((((((((.(((---...(--(((...(((((...........)))))-----.))))...)))))))(((((((....))))))))))))--)---)))..---- ( -43.00) >consensus GCUCUUCCAGUUGGCUGCCUUGGGUGGUGCGAAAUC_GGCGGCGCCAAGCCGCAAAAGGAGCCCAAAACCACGA_CGACGGCGGGUUAAGAGAUUAACAAACUGG__A___UGAAG____ ......((((((..((((((((.(((((((........(((((.....))))).......)).....)))))...))).)))))(((((....))))).))))))............... (-18.88 = -20.78 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:12 2006