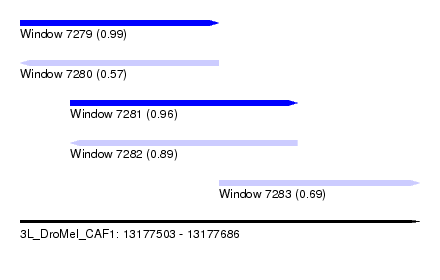
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,177,503 – 13,177,686 |
| Length | 183 |
| Max. P | 0.994682 |

| Location | 13,177,503 – 13,177,594 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
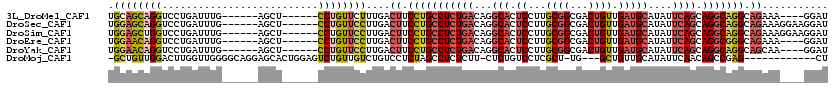
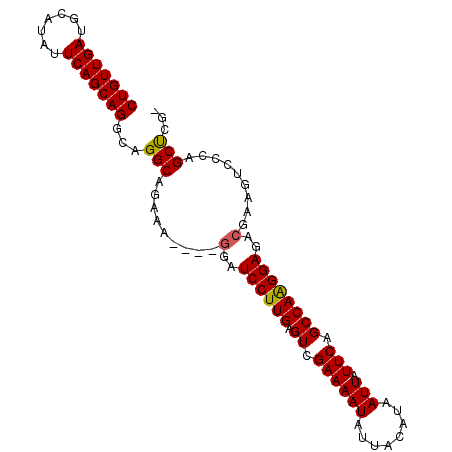
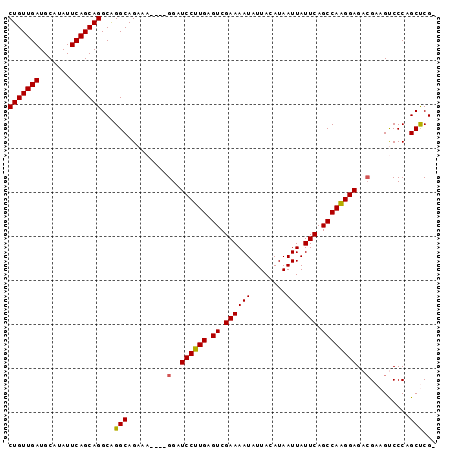
>3L_DroMel_CAF1 13177503 91 + 23771897 ACCUCCU---------GUUCCUCCUUGAGUCCUGCAGCAGGUCCUGAUUUGAGCUCCUGUUCUUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGA ...((((---------((........(((((((((((((((.........((((....))))........)))))...)).)))).))))...)))).)) ( -23.83) >DroSec_CAF1 28943 91 + 1 UCCUCCU---------GUUCCUCCUUGAGUCCUGGAGCAGGUCCUGAUUUGAGCUCCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGA ...((((---------((........(((((..((((((((..((......))..))))))))..)))))..(((((.....)))))......)))).)) ( -29.00) >DroEre_CAF1 32846 91 + 1 UCCCCCU---------GCUCCUCCUCGAGUCCUGGAACAGGUCCUGAUUUGAGCUCCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGA ....(((---------((........(((((..((((((((..((......))..))))))))..)))))..(((((.....)))))......)))).). ( -30.80) >DroYak_CAF1 27994 100 + 1 UCCUGCUCCUUCUUCUUCUACUCCUUGAGUCCUGGAACAGGUCCUGAUUUGAGCUCCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGA ((((((....................(((((..((((((((..((......))..))))))))..)))))..(((((.....)))))......)))).)) ( -31.20) >consensus UCCUCCU_________GCUCCUCCUUGAGUCCUGGAACAGGUCCUGAUUUGAGCUCCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGA ....................(.((..(((((..((((((((..((......))..))))))))..)))))..(((((.....)))))........)).). (-26.56 = -26.38 + -0.19)


| Location | 13,177,503 – 13,177,594 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -25.15 |
| Energy contribution | -26.15 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13177503 91 - 23771897 UCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAAGAACAGGAGCUCAAAUCAGGACCUGCUGCAGGACUCAAGGAGGAAC---------AGGAGGU ...((.....((((.((((((...(((.....)))...).))))).))))......))((((.(((.....(((...)))...)---------)).)))) ( -25.10) >DroSec_CAF1 28943 91 - 1 UCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGGAGCUCAAAUCAGGACCUGCUCCAGGACUCAAGGAGGAAC---------AGGAGGA ...((....))....(((((.....))))).((((..(((.((((..((......))..)))).)))..))))...........---------....... ( -25.70) >DroEre_CAF1 32846 91 - 1 UCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGGAGCUCAAAUCAGGACCUGUUCCAGGACUCGAGGAGGAGC---------AGGGGGA ((.((((........(((((.....))))).((((..((((((((..((......))..))))))))..)))).........))---------.)).)). ( -31.80) >DroYak_CAF1 27994 100 - 1 UCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGGAGCUCAAAUCAGGACCUGUUCCAGGACUCAAGGAGUAGAAGAAGAAGGAGCAGGA ...((.....((((.((((((...(((.....)))...).))))).))))......)).((((((((...((((...))))..........)))))))). ( -30.42) >consensus UCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGGAGCUCAAAUCAGGACCUGCUCCAGGACUCAAGGAGGAAC_________AGGAGGA ...((....))....(((((.....))))).((((..((((((((..((......))..))))))))..))))........................... (-25.15 = -26.15 + 1.00)

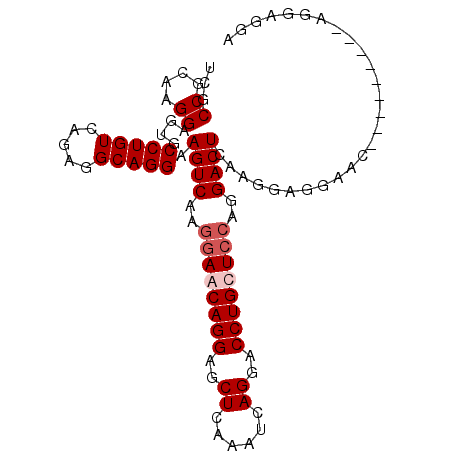
| Location | 13,177,526 – 13,177,630 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -23.00 |
| Energy contribution | -24.07 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13177526 104 + 23771897 UGCAGCAGGUCCUGAUUUG------AGCU------CCUGUUCUUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAA----GGAU ....(((((..((......------))..------)))))(((((..((.(((((((((((...(((.((...((((...)))).)))))....)))).))))))).)).))----))). ( -34.60) >DroSec_CAF1 28966 108 + 1 UGGAGCAGGUCCUGAUUUG------AGCU------CCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAAGGAAGGAU .((((((((..((......------))..------))))))))....((((((((((.....)))).((.(((((.....(((((((.......)))))))))))).))..))))))... ( -42.00) >DroSim_CAF1 27273 108 + 1 UGGAGCUGGUCCUGAUUUG------AGCU------CCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAAGGAAGGAU .((((((.(........).------))))------))(.((((((..((.(((((((((((...(((.((...((((...)))).)))))....)))).))))))).)).)))))).).. ( -42.00) >DroEre_CAF1 32869 104 + 1 UGGAACAGGUCCUGAUUUG------AGCU------CCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCGGGCAGAAA----GGAU .((((((((..((......------))..------))))))))....(((.(((((.(((.((((.....)))))))((.(((((((.......))))))).))))))).))----)... ( -39.90) >DroYak_CAF1 28026 104 + 1 UGGAACAGGUCCUGAUUUG------AGCU------CCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCAGGCAGCAA----GGAU .((((((((..((......------))..------)))))))).........(((((.....))))).(((((((.((..(((((((.......)))))))....)).))))----))). ( -42.70) >DroMoj_CAF1 77599 102 + 1 -GCUGUUGGACUUGGUUGGGGCAGGAGCACUGGAGUCUGUUGUCUGUCCUCUAGCCUCUCUU-CUCUGUCCUCGCU-UG---GCUGUUGCAUAUUCAACAGCCGAG------------CU -......((((..((..(((((.((((.((.(((........))))).)))).)))))....-))..))))..(((-((---(((((((......)))))))))))------------). ( -43.20) >consensus UGGAGCAGGUCCUGAUUUG______AGCU______CCUGUUCCUUGACUUCCUGCCUCUGACAGGCACUCCUUGCGGCGACUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAA____GGAU .((((((((..........................))))))))....((.(((((((((((...(((.((...((((...)))).)))))....)))).))))))).))........... (-23.00 = -24.07 + 1.07)

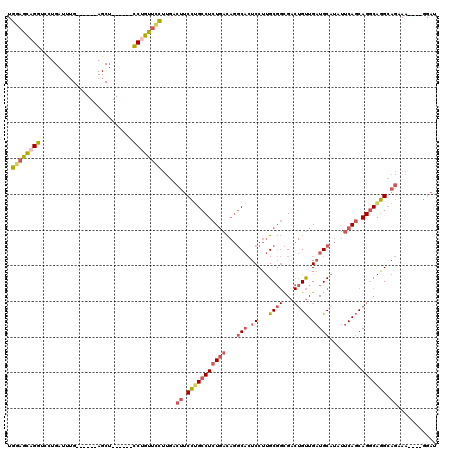
| Location | 13,177,526 – 13,177,630 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -17.33 |
| Energy contribution | -19.52 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13177526 104 - 23771897 AUCC----UUUCUGCCUGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAAGAACAGG------AGCU------CAAAUCAGGACCUGCUGCA ...(----(((((.(((((((.((((....((((.........((....)).))))...))))))))))).))..))))..(((.------((.(------(.......)).)).))).. ( -31.90) >DroSec_CAF1 28966 108 - 1 AUCCUUCCUUUCUGCCUGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGG------AGCU------CAAAUCAGGACCUGCUCCA ....((((((.((.(((((((.((((....((((.........((....)).))))...))))))))))).))..))))))..((------(((.------.............))))). ( -40.34) >DroSim_CAF1 27273 108 - 1 AUCCUUCCUUUCUGCCUGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGG------AGCU------CAAAUCAGGACCAGCUCCA ....((((((.((.(((((((.((((....((((.........((....)).))))...))))))))))).))..))))))..((------((((------............)))))). ( -40.70) >DroEre_CAF1 32869 104 - 1 AUCC----UUUCUGCCCGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGG------AGCU------CAAAUCAGGACCUGUUCCA ...(----(((((((((...(((.......)))...((((.(.((....)).)...))))..).)))))))))....((((((((------..((------......))..)))))))). ( -37.40) >DroYak_CAF1 28026 104 - 1 AUCC----UUGCUGCCUGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGG------AGCU------CAAAUCAGGACCUGUUCCA ....----(((((.(((((((.((((....((((.........((....)).))))...))))))))))).)).)))((((((((------..((------......))..)))))))). ( -41.80) >DroMoj_CAF1 77599 102 - 1 AG------------CUCGGCUGUUGAAUAUGCAACAGC---CA-AGCGAGGACAGAG-AAGAGAGGCUAGAGGACAGACAACAGACUCCAGUGCUCCUGCCCCAACCAAGUCCAACAGC- .(------------((.((((((((......)))))))---).-)))..((((...(-..(.(.(((..(((.((.((........))..)).)))..)))))..)...))))......- ( -33.60) >consensus AUCC____UUUCUGCCUGCCUGCUGAAUAUGCAUCAACAGUCGCCGCAAGGAGUGCCUGUCAGAGGCAGGAAGUCAAGGAACAGG______AGCU______CAAAUCAGGACCUGCUCCA ..............(((((((.((((....((((.........((....)).))))...))))))))))).......(((.(((............................))).))). (-17.33 = -19.52 + 2.20)

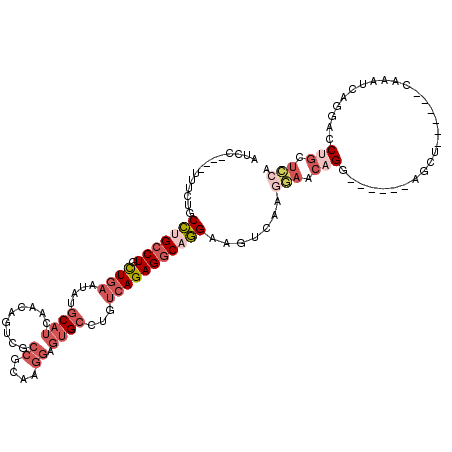
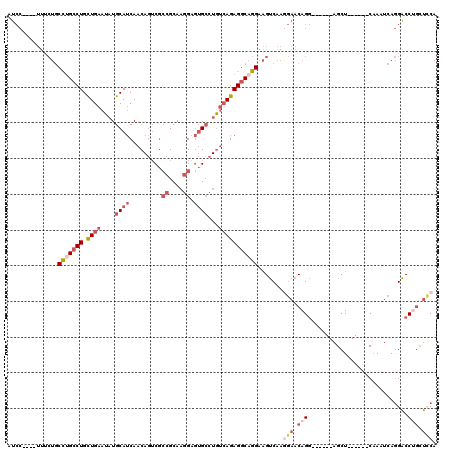
| Location | 13,177,594 – 13,177,686 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -21.11 |
| Energy contribution | -20.91 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13177594 92 + 23771897 CUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAA----GGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAAGGAGACGAAGUCCCAGCUCG- (((((((.......)))))))((.((......----(..((((((.((.((((((........))).))).))))))))..)......)).))...- ( -23.60) >DroSec_CAF1 29034 96 + 1 CUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAAGGAAGGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAAGGAGACGAAGUCCCAGCCCG- (((((((.......)))))))...(((.....(((.(..((((((.((.((((((........))).))).))))))))..)....)))..)))..- ( -26.30) >DroSim_CAF1 27341 96 + 1 CUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAAGGAAGGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAAGGAGACGAAGUCCCAGCCCG- (((((((.......)))))))...(((.....(((.(..((((((.((.((((((........))).))).))))))))..)....)))..)))..- ( -26.30) >DroEre_CAF1 32937 92 + 1 CUGUUGAUGCAUAUUCAGCAGGCGGGCAGAAA----GGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAGGGAGAGGAAGCCCCAGCUCG- (((((((.......))))))).(((((.....----...((((((.((.((((((........))).))).))))))))..((.....)).)))))- ( -25.60) >DroYak_CAF1 28094 93 + 1 CUGUUGAUGCAUAUUCAGCAGGCAGGCAGCAA----GGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAAGGAGACGAAGCCCCAGCUCGC (((((((.......)))))))((.((..((..----(..((((((.((.((((((........))).))).))))))))..)...)).)).)).... ( -25.20) >consensus CUGUUGAUGCAUAUUCAGCAGGCAGGCAGAAA____GGAUCCUUGAGUCGAAAAUAUUACAUAAUUAUUCAGCCAAGGAGACGAAGUCCCAGCUCG_ (((((((.......)))))))...(((.........(..((((((.((.((((((........))).))).))))))))..).........)))... (-21.11 = -20.91 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:11 2006