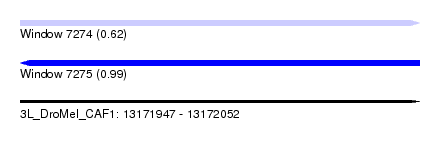
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,171,947 – 13,172,052 |
| Length | 105 |
| Max. P | 0.991354 |

| Location | 13,171,947 – 13,172,052 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -23.98 |
| Energy contribution | -23.62 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13171947 105 + 23771897 AGUGCUUGUGUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGACUGGAAUCUGGUUUUGGAUCG------------GGAUCGGGGGAUUGC ..(((((.........)))))...(((..(((((...(.(((..(((((..(((....)))((((....)))).)))))..))).)....------------.).))))..)))... ( -27.70) >DroSec_CAF1 23537 105 + 1 AGUGCUUGUGUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGACUGGAAUCUGGUUUCGGAUCG------------GGAUUGGGGGAUUGC ..(((((.........))))).......((((((........)))))).(((.........((((....))))..((..((..((...))------------..))..)).)))... ( -27.90) >DroSim_CAF1 21699 105 + 1 AGUGCUUGUGUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGACUGGAAUCUGGUUUCGGAUCG------------GGAUCGGGGGAUUGC ..(((((.........))))).......((((((........)))))).(((.........((((....))))..((((((..((...))------------..)))))).)))... ( -29.40) >DroEre_CAF1 26594 110 + 1 AGUGGUUGUGUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGAC-GGAAUCUGGUUUUGGAUCG------GGAUCUGGAUCGGGGGAUUGC .(..((((((((((.....)))))))..((((((........)))))).............((..((..(-(((.(((((((...)))))------)).))))..))..)))))..) ( -27.70) >DroYak_CAF1 22306 117 + 1 AGUGGUUGUAUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGACUGGAAUCUGGUUUUGGAUCGGCAACGGGAUAGGGAUCGGGGGAUUGC .(..((((((((((.....)))))))..((((((........)))))).............((((....))))..(((((((((...(((....)))....))))))))).)))..) ( -33.40) >consensus AGUGCUUGUGUUUGGAAAGCAAAUAUUUUCUGACAAGUGGAAGUCAGAUGUCAUAUAUAUGCCAGGGGACUGGAAUCUGGUUUUGGAUCG____________GGAUCGGGGGAUUGC .(((...(((((((.....)))))))..((((((........))))))...))).......((((....))))(((((....((.((((..............)))).))))))).. (-23.98 = -23.62 + -0.36)



| Location | 13,171,947 – 13,172,052 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -16.96 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13171947 105 - 23771897 GCAAUCCCCCGAUCC------------CGAUCCAAAACCAGAUUCCAGUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAACACAAGCACU ((........(((..------------.((((........))))...)))....((((...((((....((((((........))))))..)))).))))............))... ( -16.60) >DroSec_CAF1 23537 105 - 1 GCAAUCCCCCAAUCC------------CGAUCCGAAACCAGAUUCCAGUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAACACAAGCACU ((.(((.........------------.)))..((((.(((((.((((....)))).............((((((........))))))....))))).)))).........))... ( -16.30) >DroSim_CAF1 21699 105 - 1 GCAAUCCCCCGAUCC------------CGAUCCGAAACCAGAUUCCAGUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAACACAAGCACU ((........(((..------------..))).((((.(((((.((((....)))).............((((((........))))))....))))).)))).........))... ( -18.70) >DroEre_CAF1 26594 110 - 1 GCAAUCCCCCGAUCCAGAUCC------CGAUCCAAAACCAGAUUCC-GUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAACACAACCACU ((((......((((.......------.)))).....((((.....-.....)))).............((((((........))))))......)))).................. ( -16.80) >DroYak_CAF1 22306 117 - 1 GCAAUCCCCCGAUCCCUAUCCCGUUGCCGAUCCAAAACCAGAUUCCAGUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAAUACAACCACU ((((......((((..((......))..)))).........((.((((....)))).))..........((((((........))))))......)))).................. ( -16.40) >consensus GCAAUCCCCCGAUCC____________CGAUCCAAAACCAGAUUCCAGUCCCCUGGCAUAUAUAUGACAUCUGACUUCCACUUGUCAGAAAAUAUUUGCUUUCCAAACACAAGCACU ((((.................................((((...........)))).............((((((........))))))......)))).................. (-14.80 = -14.80 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:04 2006