
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,167,843 – 13,168,015 |
| Length | 172 |
| Max. P | 0.936168 |

| Location | 13,167,843 – 13,167,935 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 98.91 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -29.10 |
| Energy contribution | -28.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13167843 92 + 23771897 AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUUUGCC .(((.((((((((((..(((.........)))..)..))))))))))))...(((((((....))))..(((((((...)))))))..))). ( -29.40) >DroSec_CAF1 19403 92 + 1 AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUUUGCC .(((.((((((((((..(((.........)))..)..))))))))))))...(((((((....))))..(((((((...)))))))..))). ( -29.40) >DroSim_CAF1 17632 92 + 1 AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUUUGCC .(((.((((((((((..(((.........)))..)..))))))))))))...(((((((....))))..(((((((...)))))))..))). ( -29.40) >DroEre_CAF1 22736 92 + 1 AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUCUGCC .(((.((((((((((..(((.........)))..)..))))))))))))...(((((((....)))...(((((((...))))))).)))). ( -29.90) >DroYak_CAF1 18132 92 + 1 AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUCUGUC .(((.((((((((((..(((.........)))..)..))))))))))))...........((((((...(((((((...))))))))))))) ( -30.00) >consensus AGCGCGGCAUCGUUUUGAUUUGUUUGGUUAGUUGAUAAAUGAUGUCUGCACCGCAGCCUAGACAGGUCUGCCUUGGAACCCAAGGCUUUGCC .(((.((((((((((..(((.........)))..)..))))))))))))...(((((((....))))..(((((((...)))))))..))). (-29.10 = -28.94 + -0.16)


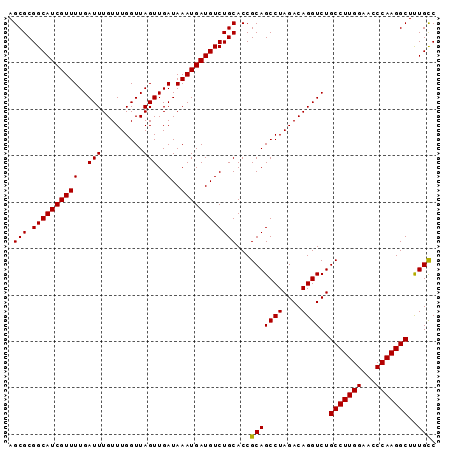
| Location | 13,167,856 – 13,167,975 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.98 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13167856 119 - 23771897 AAACCCAUCCCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ........................((((..(((.....)))((...(((((((...)))))))((.(((....)))))))))))................................... ( -23.30) >DroSec_CAF1 19416 119 - 1 AAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ...((...(((........(((((.((...(((.....))).....)).))))).......((((.(((....)))))))))))).................................. ( -24.30) >DroSim_CAF1 17645 119 - 1 AAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ...((...(((........(((((.((...(((.....))).....)).))))).......((((.(((....)))))))))))).................................. ( -24.30) >DroEre_CAF1 22749 119 - 1 AAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAGAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ........................((((..(((.....)))((((.(((((((...)))))))(((....)))...))))))))................................... ( -25.80) >DroYak_CAF1 18145 119 - 1 AAAGCCAUCCCAAUCAACCCCCAACGCCAACCAAUUACGGGACAGAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ..((((........................((......))(((((.(((((((...)))))))....)))))..))))..((((....))))........................... ( -26.40) >consensus AAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGCGGUGCAGACAUCAUUUAUCAACUAACCAAACAAAUCAAA ..((((.............(((((.((...(((.....))).....)).))))).....(((((....))))).))))..((((....))))........................... (-22.06 = -22.46 + 0.40)

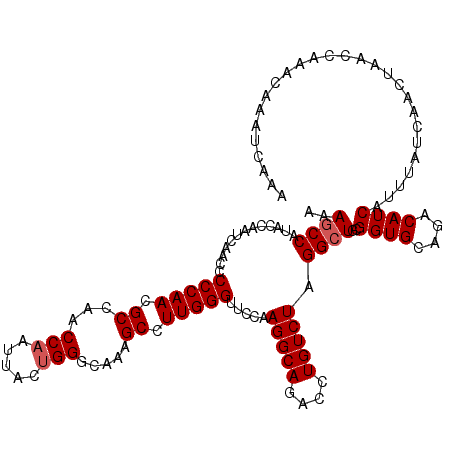
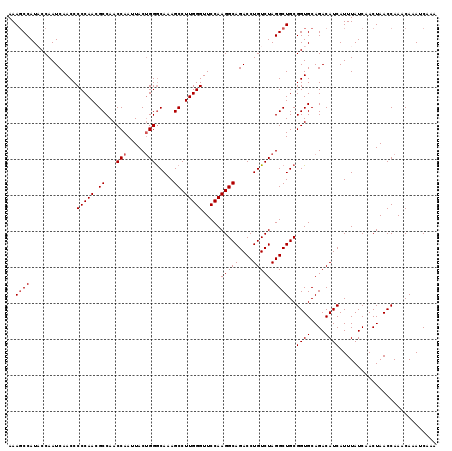
| Location | 13,167,895 – 13,168,015 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.39 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13167895 120 - 23771897 CCCAAUGGAAAAUGGAAAACCCUGGGGGCAACGUCAACCCAAACCCAUCCCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC .(((.(....).))).......((((((.............................))))))..(((..(((.....))).....(((((((...)))))))(((....))).)))... ( -30.95) >DroSec_CAF1 19455 120 - 1 CCCAAUGGAAAAUGGAAAAACCUGGGGGCAACGUCAACCCAAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC .....(((((.((((.......(((.(....).)))........))))...........(((((.((...(((.....))).....)).))))))))))..((((.(((....))))))) ( -31.16) >DroSim_CAF1 17684 120 - 1 CCCAAUGGAAAAUGGAAAACCCUGGGGGCAACGUCAACCCAAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC .....(((((.((((.......(((.(....).)))........))))...........(((((.((...(((.....))).....)).))))))))))..((((.(((....))))))) ( -31.16) >DroEre_CAF1 22788 120 - 1 CCCAGUGGAAAAUGGAAAACCCUGGGGGCGUCCUCAACCCAAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAGAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC ..((((((.....((.....))((((((.............................)))))).......)).....((((((((.(((((((...)))))))....)))))))))))). ( -36.25) >DroYak_CAF1 18184 120 - 1 CCCAAUGGAAAAUGGAAAACCCUGGGGGCAUAGUCAACCCAAAGCCAUCCCAAUCAACCCCCAACGCCAACCAAUUACGGGACAGAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC .(((.(....).))).......((((((.............................))))))..(((..((......))(((((.(((((((...)))))))....)))))..)))... ( -34.35) >consensus CCCAAUGGAAAAUGGAAAACCCUGGGGGCAACGUCAACCCAAAGCCAUACCAAUCAACCCCCAACGCCAACCAAUUACUGGGCAAAGCCUUGGGUUCCAAGGCAGACCUGUCUAGGCUGC .(((.(....).))).......((((((.............................))))))..(((..(((.....))).....(((((((...)))))))(((....))).)))... (-30.19 = -30.39 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:00 2006