
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,160,258 – 13,160,352 |
| Length | 94 |
| Max. P | 0.916066 |

| Location | 13,160,258 – 13,160,352 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
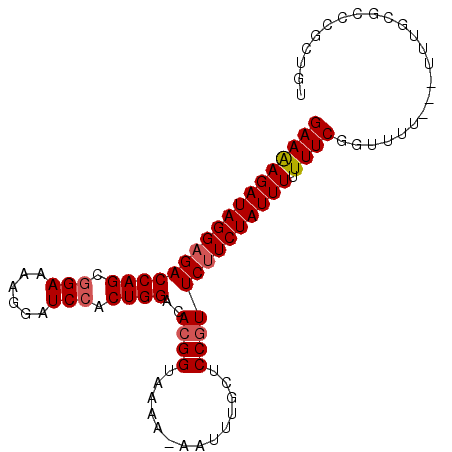
>3L_DroMel_CAF1 13160258 94 + 23771897 GAAAAGAUAGGAGACCAGCGGAAAAGGAUCCACUGGACACGGUAAAA-AAAUUGCUCCGUUCUUCUAUUUUUUUCGGUUUU---UUUGCGCCCGCUGU ((((((((((((((((((.(((......))).))))..((((.....-........)))))))))))))))))).(((...---.....)))...... ( -26.92) >DroSec_CAF1 11867 97 + 1 GAAAAGAUAGGAGACCAGCAGAAAAGGAUCCACUGGACACGGUAAAA-AAUUUGCUCCGUUCUUCUAUUUUUUUCGGUUUUUUUUUUGCGCCCGCUGU ((((((((((((((((((..((......))..))))..((((.....-........)))))))))))))))))).............((....))... ( -21.82) >DroSim_CAF1 10092 96 + 1 GAAAAGAUAGGAGACCAGCGGAAAAGGAUCCACUGGACACGGUAAAA-AAUUUGCUCCGUUCUUCUAUUUUUUUCGGUUUU-UUUUUGCGCCCGCUGU ((((((((((((((((((.(((......))).))))..((((.....-........)))))))))))))))))).......-.....((....))... ( -26.52) >DroEre_CAF1 15251 93 + 1 GAAGAGAUAGGAGACCAGCGGAAAAGGAUCCACUGGACACGGUAAAAAAAUGUGCUCCUUUCCUCUAUUU-UUUCGGUUU----UUUGCGACCGCUGU ((((((((((..((((((.(((......))).)))).((((.........))))......))..))))))-))))((((.----.....))))..... ( -27.50) >DroYak_CAF1 10296 92 + 1 GAAAAGAUAGGAGACCAGCGGAAAAGGAUCCACUGGACACGGUAAAA-AAUUUGCUCCGUUCUUCUAUUU-UUUCGGUUU----UUUGCGCCCCCUGU ((((((((((((((((((.(((......))).))))..((((.....-........))))))))))))))-))))(((..----.....)))...... ( -29.82) >consensus GAAAAGAUAGGAGACCAGCGGAAAAGGAUCCACUGGACACGGUAAAA_AAUUUGCUCCGUUCUUCUAUUUUUUUCGGUUUU___UUUGCGCCCGCUGU ((((((((((((((((((.(((......))).))))..((((..............)))))))))))))).))))....................... (-21.86 = -22.30 + 0.44)

| Location | 13,160,258 – 13,160,352 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
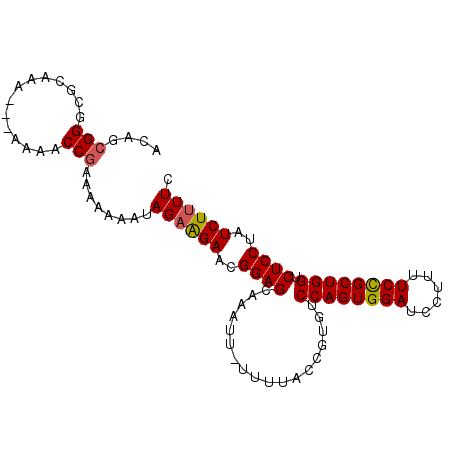
>3L_DroMel_CAF1 13160258 94 - 23771897 ACAGCGGGCGCAAA---AAAACCGAAAAAAAUAGAAGAACGGAGCAAUUU-UUUUACCGUGUCCAGUGGAUCCUUUUCCGCUGGUCUCCUAUCUUUUC ....(((.......---....))).((((.((((.(((((((........-.....))))..((((((((......))))))))))).)))).)))). ( -23.72) >DroSec_CAF1 11867 97 - 1 ACAGCGGGCGCAAAAAAAAAACCGAAAAAAAUAGAAGAACGGAGCAAAUU-UUUUACCGUGUCCAGUGGAUCCUUUUCUGCUGGUCUCCUAUCUUUUC ....(((..............))).((((.((((.(((((((........-.....))))..((((..((......))..))))))).)))).)))). ( -20.66) >DroSim_CAF1 10092 96 - 1 ACAGCGGGCGCAAAAA-AAAACCGAAAAAAAUAGAAGAACGGAGCAAAUU-UUUUACCGUGUCCAGUGGAUCCUUUUCCGCUGGUCUCCUAUCUUUUC ....(((.........-....))).((((.((((.(((((((........-.....))))..((((((((......))))))))))).)))).)))). ( -23.54) >DroEre_CAF1 15251 93 - 1 ACAGCGGUCGCAAA----AAACCGAAA-AAAUAGAGGAAAGGAGCACAUUUUUUUACCGUGUCCAGUGGAUCCUUUUCCGCUGGUCUCCUAUCUCUUC ....((((......----..))))...-....(((((..((((((((...........))).((((((((......)))))))).))))).))))).. ( -27.10) >DroYak_CAF1 10296 92 - 1 ACAGGGGGCGCAAA----AAACCGAAA-AAAUAGAAGAACGGAGCAAAUU-UUUUACCGUGUCCAGUGGAUCCUUUUCCGCUGGUCUCCUAUCUUUUC ..(((((((.....----.........-.....((...((((........-.....)))).))(((((((......))))))))))))))........ ( -25.42) >consensus ACAGCGGGCGCAAA___AAAACCGAAAAAAAUAGAAGAACGGAGCAAAUU_UUUUACCGUGUCCAGUGGAUCCUUUUCCGCUGGUCUCCUAUCUUUUC ....(((..............)))........((((((..((((..................((((((((......)))))))).))))..)))))). (-20.15 = -20.23 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:48 2006