| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,334,121 – 1,334,234 |
| Length | 113 |
| Max. P | 0.838328 |

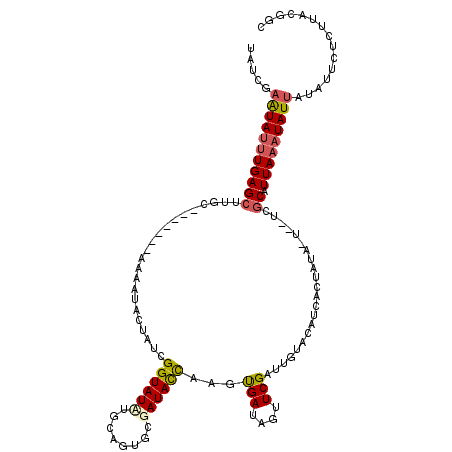
| Location | 1,334,121 – 1,334,234 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1334121 113 + 23771897 UAUCGAAUAUUUGAGCUUGC-------GAAAUACUAUCGGUAUUUGCAGUGCGAUACCGAGUGAUAGUUCGAUUGUACAUCACUAUAUUGAUCGCAUUACAUAUUAUAUUCUCUUAUGGU ....((((((.((.....((-------.((((((.....))))))))((((((((....((((((.((........))))))))......)))))))).))....))))))......... ( -26.90) >DroSec_CAF1 6286 98 + 1 UAUCGAAUAUUUGAGCUUGC-------AAAAUACUAUCGGUAUAUGCAGUGCGAUACCAAGUGAUAGUUCGAUUGU---------------UCGCAUUAAAUAUUAUAUUCCCUUACGGC .....(((((((((((..((-------((...(((((((((((.(((...))))))))....))))))....))))---------------..)).)))))))))............... ( -21.70) >DroSim_CAF1 6323 98 + 1 UAUCGAAUAUUUGAGCUUGC-------AAAAUACUAUCGGUAUAUGCAGUGCGAUACCAAGUGAUAGUUCGAUUGU---------------UCGCAUUAAAUAUUAUAUUCGCUUACGGC .....(((((((((((..((-------((...(((((((((((.(((...))))))))....))))))....))))---------------..)).)))))))))............... ( -21.70) >DroEre_CAF1 6462 110 + 1 UAUCGAGUAUUUGAGCUUGCUAGUGAAAACACAUUAUCGGUAUUUUCAGAUCGAUAUCAAUCGAUAGCUCGAUUGCACAUCACUAUAAUAAUCGCAUUAAUUAUCG--AUAU-------- ((((((....(((((((((.((((((........(((((((........))))))).(((((((....)))))))....))))))))).....)).))))...)))--))).-------- ( -25.30) >DroYak_CAF1 6480 111 + 1 UAUCGAAUAUUUGAGCUUGC-------AAAACACUAUCGGUAUCUUCGGUCCGAUACUGAGCGAUAGCUCGAUUGCACAUCACUAUAUUAUUGCCAUUAAAUAUUA--UUCUUUUAUGGU .....(((((.(((...(((-------((.....((((((...(....).)))))).(((((....))))).)))))..)))..)))))...(((((.(((.....--...))).))))) ( -26.80) >consensus UAUCGAAUAUUUGAGCUUGC_______AAAAUACUAUCGGUAUAUGCAGUGCGAUACCAAGUGAUAGUUCGAUUGUACAUCACUAUA_U__UCGCAUUAAAUAUUAUAUUCUCUUACGGC .....(((((((((((......................((((((........))))))...(((....)))......................)).)))))))))............... ( -9.86 = -10.26 + 0.40)

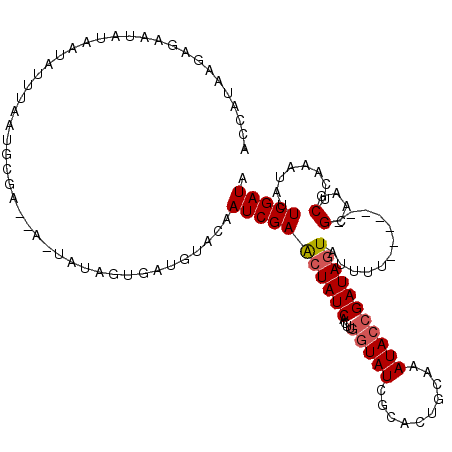
| Location | 1,334,121 – 1,334,234 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -7.42 |
| Energy contribution | -7.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1334121 113 - 23771897 ACCAUAAGAGAAUAUAAUAUGUAAUGCGAUCAAUAUAGUGAUGUACAAUCGAACUAUCACUCGGUAUCGCACUGCAAAUACCGAUAGUAUUUC-------GCAAGCUCAAAUAUUCGAUA .........((((((.....((..(((((..(((((.(((((((........)).)))))(((((((.((...))..)))))))..)))))))-------))).))....)))))).... ( -23.60) >DroSec_CAF1 6286 98 - 1 GCCGUAAGGGAAUAUAAUAUUUAAUGCGA---------------ACAAUCGAACUAUCACUUGGUAUCGCACUGCAUAUACCGAUAGUAUUUU-------GCAAGCUCAAAUAUUCGAUA .((....))((((((.........(((((---------------...(((((........))))).))))).((((.((((.....))))..)-------))).......)))))).... ( -20.10) >DroSim_CAF1 6323 98 - 1 GCCGUAAGCGAAUAUAAUAUUUAAUGCGA---------------ACAAUCGAACUAUCACUUGGUAUCGCACUGCAUAUACCGAUAGUAUUUU-------GCAAGCUCAAAUAUUCGAUA ........(((((((.........(((((---------------...(((((........))))).))))).((((.((((.....))))..)-------))).......)))))))... ( -18.70) >DroEre_CAF1 6462 110 - 1 --------AUAU--CGAUAAUUAAUGCGAUUAUUAUAGUGAUGUGCAAUCGAGCUAUCGAUUGAUAUCGAUCUGAAAAUACCGAUAAUGUGUUUUCACUAGCAAGCUCAAAUACUCGAUA --------.(((--(((.......(((((((((....))))).))))...(((((((((((....)))))).(((((((((.......)))))))))......)))))......)))))) ( -28.90) >DroYak_CAF1 6480 111 - 1 ACCAUAAAAGAA--UAAUAUUUAAUGGCAAUAAUAUAGUGAUGUGCAAUCGAGCUAUCGCUCAGUAUCGGACCGAAGAUACCGAUAGUGUUUU-------GCAAGCUCAAAUAUUCGAUA .((((.(((...--.....))).))))....(((((..(((..(((((....(((((((....(((((........))))))))))))...))-------)))...))).)))))..... ( -24.00) >consensus ACCAUAAGAGAAUAUAAUAUUUAAUGCGA__A_UAUAGUGAUGUACAAUCGAACUAUCACUUGGUAUCGCACUGCAAAUACCGAUAGUAUUUU_______GCAAGCUCAAAUAUUCGAUA ...............................................(((((((((((....(((((..........)))))))))))............(....)........))))). ( -7.42 = -7.78 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:29 2006