
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,158,396 – 13,158,508 |
| Length | 112 |
| Max. P | 0.570264 |

| Location | 13,158,396 – 13,158,508 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -29.60 |
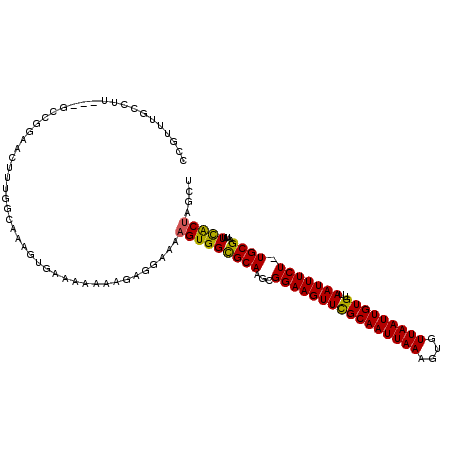
| Consensus MFE | -18.62 |
| Energy contribution | -18.15 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
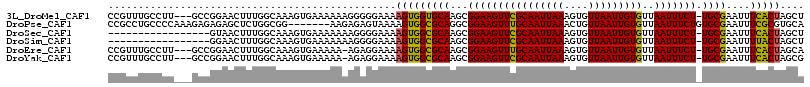
>3L_DroMel_CAF1 13158396 112 - 23771897 CCGUUUGCCUU---GCCGGAACUUUGGCAAAGUGAAAAAAGGGGGAAAAGUGGUGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCU ((.((..(.((---(((((....))))))).)..))....))......(((((.....((((((((((((((((((....)))))))))..)))))).-))).....))))).... ( -31.60) >DroPse_CAF1 14840 109 - 1 CCGCCUGCCCCAAAGAGAGAGCUCUGGCGG-------AAGAGAGUAAAAGUGGCGCAGGCGGAAGUUUGCAAUUAAACUGUUAAUUGUGUUAAUUUCUGUGCGAAUUUCGCGUGCA .(((((((.(((........(((((..(..-------..)))))).....))).)))))))((((((.((((((((....))))))))...)))))).(..((.......))..). ( -36.82) >DroSec_CAF1 10030 98 - 1 -----------------GUAACUUUGGCAAAGUGAAAAAAAGGGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCU -----------------........(((..(((((((...............((....))((((((((((((((((....)))))))))..)))))))-......))))))).))) ( -24.00) >DroSim_CAF1 8240 98 - 1 -----------------GGAACUUUGGCAAAGUGAAAAAAAGGGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUUACUAGCU -----------------........(((..(((((((...............((....))((((((((((((((((....)))))))))..)))))))-......))))))).))) ( -21.80) >DroEre_CAF1 11956 111 - 1 CCGUUUGCCUU---GCCGGAACUUUGGCAAAGUGAAAAA-AGAGGAAAAGUGGCGCAAGCGGAAGUUUGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCA .....(((.((---(((((....)))))))(((((((..-.(((((((..((((((((((....))))))((((((....))))))..)))).)))))-).)...))))))).))) ( -32.70) >DroYak_CAF1 8527 111 - 1 CCGUUUGCCUU---GCCGGAACUUUGGCAAAGUGAAAAA-AGAGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU-UGCGAAUUUCACUAGCG ......((.((---(((((....)))))))(((((((..-............((....))((((((((((((((((....)))))))))..)))))))-......))))))).)). ( -30.70) >consensus CCGUUUGCCUU___GCCGGAACUUUGGCAAAGUGAAAAAAAGAGGAAAAGUGGCGCAAGCGGAAGUUCGCAAUUAAAGUGUUAAUUGUGUUAAUUUCU_UGCGAAUUUCACUAGCU ................................................(((((((((...((((((((((((((((....)))))))))..))))))).))))....))))).... (-18.62 = -18.15 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:44 2006