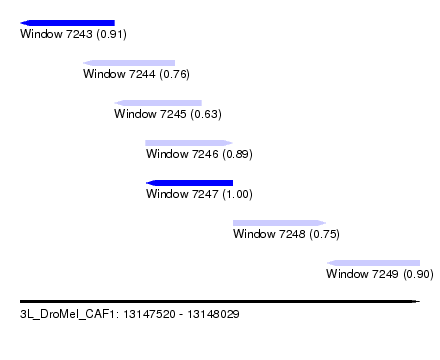
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,147,520 – 13,148,029 |
| Length | 509 |
| Max. P | 0.996479 |

| Location | 13,147,520 – 13,147,640 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -47.63 |
| Consensus MFE | -44.75 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147520 120 - 23771897 CACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCCCGAGUCCCAGAGGCCGCACUUCAUCAACUUGGGGUUCGGUCUCGAAUGUGCUAAAAAAUUGAUUGAUCCUGCAGGAGGCG ..(((.(((((.(((((.(((((((((((((((........))))))).(((((((.((((((......)))))).)))))))..........)))))))).)))))..))))))))... ( -48.70) >DroSec_CAF1 135176 120 - 1 CACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCCCGAGUCCCAGAGGCCGCACUUCAUCAACUUGGGGUUCGGUCUCGAAUGUGCUAAAAAAUUGAUUGAUCCUGCAGGAGGCG ..(((.(((((.(((((.(((((((((((((((........))))))).(((((((.((((((......)))))).)))))))..........)))))))).)))))..))))))))... ( -48.70) >DroSim_CAF1 135047 120 - 1 CACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCCCGAGUCCCAGAGGCCGCACUUCAUCAACUUGGGGUUCGGUCUCGAAUGUGCUAAAAAAUUGAUUGAUCCUGCAGGAGGCG ..(((.(((((.(((((.(((((((((((((((........))))))).(((((((.((((((......)))))).)))))))..........)))))))).)))))..))))))))... ( -48.70) >DroYak_CAF1 136761 120 - 1 CACUCGCUGCAUAUUAAACAGUUUUUGAGAUUCAGUCACCCGAGUCCCAGAGGCCGCACUUCAUCAACUUGGGGCUCGGGCUCGAAUGUGCUAAAAAAUUGAUUGAUCCUGCAGGAGGCG ..(((.(((((.(((((.((((((((.((...((.((.(((((((((((((((.....)))).......)))))))))))...)).))..)).)))))))).)))))..))))))))... ( -44.41) >consensus CACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCCCGAGUCCCAGAGGCCGCACUUCAUCAACUUGGGGUUCGGUCUCGAAUGUGCUAAAAAAUUGAUUGAUCCUGCAGGAGGCG ..(((.(((((.(((((.(((((((((((((((........))))))).(((((((.((((((......)))))).)))))))..........)))))))).)))))..))))))))... (-44.75 = -45.50 + 0.75)

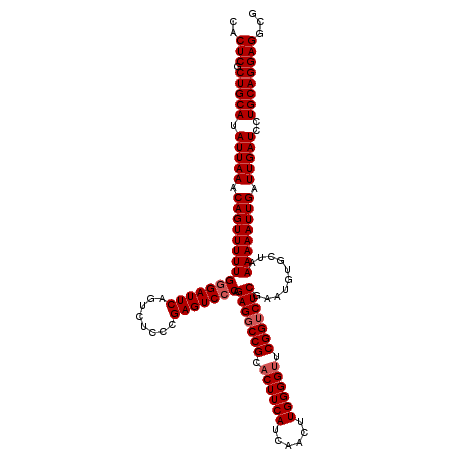

| Location | 13,147,600 – 13,147,717 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -38.24 |
| Energy contribution | -37.92 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147600 117 - 23771897 AGGAGGUGGAGACACCUCUUC---CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGUCACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCC ((((((((....)))))))).---....((((.(.(((((((((((((((((......((..(((((....)))))..)).....)))))))......)))))))..))).).))))... ( -39.20) >DroSec_CAF1 135256 117 - 1 AGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGAAAUUGGUGCGGUUGCUCAAUGUCGUCGCGCUCGGCGGUGUCACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCC ((((((((....)))))))).---....((((.(.((((((((((((((((..((.....))..))))))...((((((.....))))))........)))))))..))).).))))... ( -46.30) >DroSim_CAF1 135127 117 - 1 AGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGUCACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCC ((((((((....)))))))).---....((((.(.(((((((((((((((((......((..(((((....)))))..)).....)))))))......)))))))..))).).))))... ( -41.90) >DroYak_CAF1 136841 120 - 1 AGAAGGUGGAGGCACCUCCUUCCCUUCUGGUUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGUCACUCGCUGCAUAUUAAACAGUUUUUGAGAUUCAGUCACC ((((((.((((....))))...))))))......(((.((((((((((((((......((..(((((....)))))..)).....)))))))......))))))).)))........... ( -37.80) >consensus AGGAGGUGGAGACACCUCCUC___CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGUCACUCGCUGCAUAUUAAACAGUUUUUGGGAUUCAGUCUCC ((((((((....))))))))......((((....(((.((((((((((((((......((..(((((....)))))..)).....)))))))......))))))).)))..))))..... (-38.24 = -37.92 + -0.31)



| Location | 13,147,640 – 13,147,751 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -31.66 |
| Energy contribution | -31.97 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147640 111 - 23771897 AGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCUUC---CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGU .........((((((..((((((.(((.((((------(.((((((((....)))))))).---)))))))).))).(((.(((((.((....))))))).)))....)))...)))))) ( -44.60) >DroSec_CAF1 135296 111 - 1 AGAUGGAUGGGACCGAAUGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGAAAUUGGUGCGGUUGCUCAAUGUCGUCGCGCUCGGCGGUGU ...(((((((((((((......))))).((((------(.((((((((....)))))))).---))))).)))).)))).....(((((((..((.....))..)))))))......... ( -47.80) >DroSim_CAF1 135167 111 - 1 AGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGU .........((((((..((((((.(((.((((------(.((((((((....)))))))).---)))))))).))).(((.(((((.((....))))))).)))....)))...)))))) ( -47.30) >DroYak_CAF1 136881 118 - 1 AGACGGAUGGGAUUGAAUGCAGCA--UGCCAACUGCAUGUAGAAGGUGGAGGCACCUCCUUCCCUUCUGGUUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGU .(((((((((.(((((..((....--.))(((((((((..((((((.((((....))))...))))))((......)).......))))))))).))))).))))).)).))........ ( -42.80) >consensus AGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA______GUAGGAGGUGGAGACACCUCCUC___CUCUGGCUCACUCCGAAAUUGGUGCAGUUGCUCAAUGUCGUCGCGCUCGGCGGUGU .((((((((((...((..(((.....)))...........((((((((....)))))))).....))...)))).)))((.(((((.((....))))))).)))))(((((....))))) (-31.66 = -31.97 + 0.31)

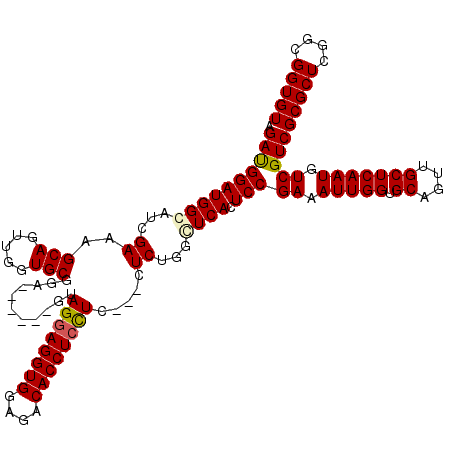

| Location | 13,147,680 – 13,147,791 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147680 111 + 23771897 UCGGAGUGAGCCAGAG---GAAGAGGUGUCUCCACCUCCUAC------UCCGCACCAACUGCUUUCGAUGCCAUCCAUCUUUUGUCGGUUGACUGCAGUUCAUAAAUUUUUAAAACGCUC ...(((((.((..(((---...((((((....))))))...)------)).))...((((((..(((..(((...((.....))..))))))..))))))...............))))) ( -30.40) >DroSec_CAF1 135336 111 + 1 UCGGAGUGAGCCAGAG---GAGGAGGUGUCUCCACCUCCUAC------UCCGCACCAACUGCAUUCGGUCCCAUCCAUCUUUUGUCGGUUGACUGCAGUUCAUAAAUUUUUAAAACGCUC ...(((((.((..(((---.((((((((....)))))))).)------)).))...(((((((.((((.((....((.....))..)))))).)))))))...............))))) ( -36.70) >DroSim_CAF1 135207 110 + 1 UCGGAGUGAGCCAGAG---GAGGAGGUGUCUCCACCUCCUAC------UCCGCACCAACUGCUUUCGAUGCCAUCCAUCUUUUGUCGGUUGACUGCAGUUCAUCCGUAU-AGGGUCGCUC ...((((((.((.(((---.((((((((....)))))))).)------)).(((.(((((((....((((.....))))....).))))))..))).............-.)).)))))) ( -41.60) >DroYak_CAF1 136921 117 + 1 UCGGAGUGAACCAGAAGGGAAGGAGGUGCCUCCACCUUCUACAUGCAGUUGGCA--UGCUGCAUUCAAUCCCAUCCGUCUUUUGUCGCUUGACUGCAGUUCAUAAAUUUUU-AAACGCUC .....((((((..((.((((((((((((....))))))))..(((((((.....--.)))))))....)))).))...........((......)).))))))........-........ ( -34.00) >consensus UCGGAGUGAGCCAGAG___GAGGAGGUGUCUCCACCUCCUAC______UCCGCACCAACUGCAUUCGAUCCCAUCCAUCUUUUGUCGGUUGACUGCAGUUCAUAAAUUUUUAAAACGCUC ...(((((............((((((((....))))))))................(((((((.((((..((...((.....))..)))))).)))))))...............))))) (-25.16 = -25.47 + 0.31)



| Location | 13,147,680 – 13,147,791 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -30.33 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147680 111 - 23771897 GAGCGUUUUAAAAAUUUAUGAACUGCAGUCAACCGACAAAAGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCUUC---CUCUGGCUCACUCCGA ((((............(((.((((((.(((....)))....((((.....))))....)))))).)))((((------(.((((((((....)))))))).---)))))))))....... ( -46.40) >DroSec_CAF1 135336 111 - 1 GAGCGUUUUAAAAAUUUAUGAACUGCAGUCAACCGACAAAAGAUGGAUGGGACCGAAUGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGA ((((............(((.((((((((((....)))......(((......)))..))))))).)))((((------(.((((((((....)))))))).---)))))))))....... ( -47.60) >DroSim_CAF1 135207 110 - 1 GAGCGACCCU-AUACGGAUGAACUGCAGUCAACCGACAAAAGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA------GUAGGAGGUGGAGACACCUCCUC---CUCUGGCUCACUCCGA (((.((.((.-...((.((.((((((.(((....)))....((((.....))))....)))))).)).))((------(.((((((((....)))))))).---))).)).)).)))... ( -49.50) >DroYak_CAF1 136921 117 - 1 GAGCGUUU-AAAAAUUUAUGAACUGCAGUCAAGCGACAAAAGACGGAUGGGAUUGAAUGCAGCA--UGCCAACUGCAUGUAGAAGGUGGAGGCACCUCCUUCCCUUCUGGUUCACUCCGA ..((((((-(........))))).)).(((....)))......((((..((((...((((((..--......)))))).(((((((.((((....))))...)))))))))))..)))). ( -36.20) >consensus GAGCGUUUUAAAAAUUUAUGAACUGCAGUCAACCGACAAAAGAUGGAUGGCAUCGAAAGCAGUUGGUGCGGA______GUAGGAGGUGGAGACACCUCCUC___CUCUGGCUCACUCCGA ((((............(((.((((((.(((....)))......(((......)))...)))))).)))((((........((((((((....)))))))).....))))))))....... (-30.33 = -31.14 + 0.81)



| Location | 13,147,791 – 13,147,910 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.99 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -33.32 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147791 119 + 23771897 GCCGGCGCGAACCAAAACAAACAAGUUUUGAAUCGCCGCCAGGAUCGGUGGAACGCAUCCCAAAAGGUG-GAGUGUGUGGACAGAAGGAGACUGGCAGACACGCAUGUAAUCAAUUUUUA ((((((((((..((((((......))))))..))).)))).((((((......)).)))).....))).-..(((((((......((....))......))))))).............. ( -38.80) >DroSec_CAF1 135447 119 + 1 GCCGCCGCGAACCAAAACAAACAAGUUUUGAAUCGCCGCCAGGAUCGGCAGAACGCAUCCCAAAAGGUG-GAGUGUGUGGACAGAAGGAGACUGGCAGACACGCAUGUAAUCAAUUUUUA .(((((((((..((((((......))))))..)))).....((((((......)).)))).....))))-).(((((((......((....))......))))))).............. ( -38.40) >DroYak_CAF1 137038 120 + 1 GCCGGCGCGAACCAAAACAAACAAGUUUUGAACCGCCGCCAGGAUCGGUGGAACGCAUCCCAAAACGCGAGUGUGUGUGGACAGAAGGAGACUGGCAGACACGCAUAUAAUCAAUUUUUA ((.((((((...((((((......))))))...)).)))).((((((......)).))))......))(((((((((((......((....))......)))))))))..))........ ( -37.00) >consensus GCCGGCGCGAACCAAAACAAACAAGUUUUGAAUCGCCGCCAGGAUCGGUGGAACGCAUCCCAAAAGGUG_GAGUGUGUGGACAGAAGGAGACUGGCAGACACGCAUGUAAUCAAUUUUUA ((((((((((..((((((......))))))..))).)))).((((((......)).)))).....)))....(((((((......((....))......))))))).............. (-33.32 = -34.10 + 0.78)

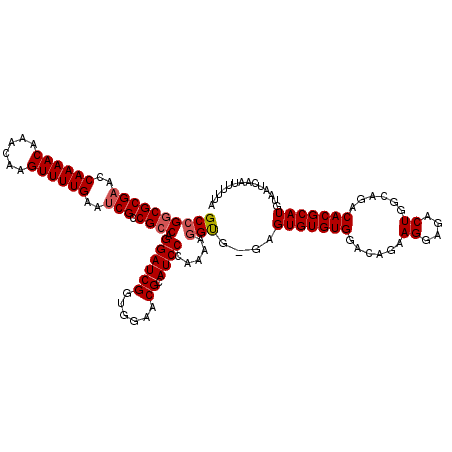
| Location | 13,147,910 – 13,148,029 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -38.68 |
| Energy contribution | -38.46 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147910 119 - 23771897 GUAGGAGAAUGCGAGAACAUUCAGUUUAUCGUUUC-CCUCGGCGGCAACGGGUAAUCCAAUUAUAAUAGAGCUGGUCCGCCGGCUGCACCUGCCUCACUUGGGGGCAUUCUUCUAAAAUA .(((((((((((((((((.....)))).))....(-(((.((.((((...((....))...........((((((....)))))).....))))...)).)))))))))))))))..... ( -40.10) >DroSec_CAF1 135566 119 - 1 GUAGGAGAAUGCGAGAACAUUCAGUUUAUCGUUUC-CCUCGGCGGCAACGGGUAAUCCAAUUAUAAUAGAGCUGGUCCGCCGGCUGCACCUGCCUCACUUGGGGGCAUUCUUCUAAAAUA .(((((((((((((((((.....)))).))....(-(((.((.((((...((....))...........((((((....)))))).....))))...)).)))))))))))))))..... ( -40.10) >DroYak_CAF1 137158 120 - 1 GUAGGAGAAUGCGAGAACAUUCAGUUUAUCGUUUCCCCUUGGCGGCAACGGGUAAUCCAAUUAUAAUAGAGCUGGUCCGCCGGCUGCACCUGCCUCACUUGGGGGCAUUCUUCUAAAAUA .(((((((((((((((((.....)))).))....((((..((.((((...((....))...........((((((....)))))).....)))).).)..)))))))))))))))..... ( -43.50) >consensus GUAGGAGAAUGCGAGAACAUUCAGUUUAUCGUUUC_CCUCGGCGGCAACGGGUAAUCCAAUUAUAAUAGAGCUGGUCCGCCGGCUGCACCUGCCUCACUUGGGGGCAUUCUUCUAAAAUA .(((((((((((((((((.....)))).))......(((((((((..((.(((.................))).)))))))(((.......)))......)))))))))))))))..... (-38.68 = -38.46 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:38 2006