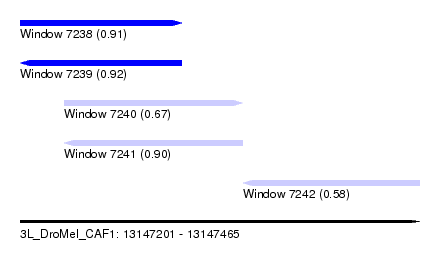
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,147,201 – 13,147,465 |
| Length | 264 |
| Max. P | 0.915371 |

| Location | 13,147,201 – 13,147,308 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147201 107 + 23771897 ----------UAUUUGUU-UGUGACAAAUAACUUUCCAUAGUUCAAG--UACUCGUUAGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUC ----------((((((.(-((((.............)))))..))))--))................(((((((((((.(((((...((((....))))..))))).)))))).))))). ( -22.92) >DroSec_CAF1 134855 109 + 1 ----------AAUUUGUU-UGUGACAAAUAACUUUCCAUAGUUCAAGCGCACUCGUUUGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUC ----------.(((((((-...)))))))...........(..((((((....))))))..).....(((((((((((.(((((...((((....))))..))))).)))))).))))). ( -26.00) >DroSim_CAF1 134728 107 + 1 ----------AAUUUAUU-UCUUACAAAGAACUUUCCAUAGUUCAAG--CACUCGUUUGCCCCAGUCAUUGCUUCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUC ----------........-.........(((((......)))))...--.....((...(((.(((...((.(((((..((((.....))))..)))))))....))).)))..)).... ( -23.50) >DroYak_CAF1 136403 118 + 1 AUGCAAAUAGAAUUUGUUAUAUGACAAGAAACUAUCCACAGUUCAAG--CACUCGUUAGCCCCAGCUAAUGCUCCCAAAUGGAACGUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUC .(((........((((((....)))))).((((......))))...)--))...((((((....))))))((((((((.(((((.(((.((....))))).))))).)))))).)).... ( -28.20) >consensus __________AAUUUGUU_UGUGACAAAUAACUUUCCAUAGUUCAAG__CACUCGUUAGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUC ............((((((....)))))).((((......))))........................(((((((((((.(((((...((((....))))..))))).)))))).))))). (-18.48 = -18.85 + 0.37)



| Location | 13,147,201 – 13,147,308 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -23.23 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147201 107 - 23771897 GAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCUAACGAGUA--CUUGAACUAUGGAAAGUUAUUUGUCACA-AACAAAUA---------- .(((((.((((((((..((((((((....)).))))))..)))))))))))))(((....))).(((((((--(((..........))).)))))))....-........---------- ( -27.00) >DroSec_CAF1 134855 109 - 1 GAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCAAACGAGUGCGCUUGAACUAUGGAAAGUUAUUUGUCACA-AACAAAUU---------- .(((((.((((((((..((((((((....)).))))))..)))))))))))))(..((.((((((((((....))))((((......)))).)))))).))-..).....---------- ( -31.40) >DroSim_CAF1 134728 107 - 1 GAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGAAGCAAUGACUGGGGCAAACGAGUG--CUUGAACUAUGGAAAGUUCUUUGUAAGA-AAUAAAUU---------- ..((((..((((.((....(((((((..((((.....))))..))).))))...)))))))))).....((--(..(((((......)))))...)))...-........---------- ( -27.30) >DroYak_CAF1 136403 118 - 1 GAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAACGUUCCAUUUGGGAGCAUUAGCUGGGGCUAACGAGUG--CUUGAACUGUGGAUAGUUUCUUGUCAUAUAACAAAUUCUAUUUGCAU ...(((.((((((((..((((((((....)).))))))..)))))))))))(((((....)))))..(((.--.....)))(..(((((....((((......))))...)))))..).. ( -34.50) >consensus GAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCAAACGAGUG__CUUGAACUAUGGAAAGUUAUUUGUCACA_AACAAAUU__________ .(((((.((((((((..((((((((....)).))))))..))))))))))))).(((.(......).)))......(((((......)))))(((((......)))))............ (-23.23 = -23.85 + 0.62)



| Location | 13,147,230 – 13,147,348 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147230 118 + 23771897 GUUCAAG--UACUCGUUAGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUCAAGGCCACCACUUCUCCAACCUGCCUAUUUAGCCACUUGU ...((((--(....(((((........(((((((((((.(((((...((((....))))..))))).)))))).)))))..((((.................))))..))))).))))). ( -26.43) >DroSec_CAF1 134884 120 + 1 GUUCAAGCGCACUCGUUUGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUCAAGGCCACCACUUCUCCAACCCGCCUAUUUAGCCACUUGU (..((((((....))))))..)..((((((((((((((.(((((...((((....))))..))))).)))))).)))))...)))................................... ( -27.10) >DroSim_CAF1 134757 118 + 1 GUUCAAG--CACUCGUUUGCCCCAGUCAUUGCUUCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUCAAGGCCACCACUUCUCCAACCCGCCUAUUUAGCCACUUGU ...((((--.....(.((((((((((...((.(((((..((((.....))))..)))))))....)).))))..)))).).((((.................)))).........)))). ( -24.53) >DroYak_CAF1 136443 118 + 1 GUUCAAG--CACUCGUUAGCCCCAGCUAAUGCUCCCAAAUGGAACGUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUCAAGGCCACCACUUCUCCAACCCGCCUAUUUGGCCACUUGU ...((((--.....((((((....))))))((((((((.(((((.(((.((....))))).))))).)))))).))......(((((......................))))).)))). ( -31.05) >consensus GUUCAAG__CACUCGUUAGCCCCAGCCAUUGCUCCCAAAUGGAACAUUUCCAAAUGGAACAUUUUACUUGGGAUGCAAUCAAGGCCACCACUUCUCCAACCCGCCUAUUUAGCCACUUGU ...((((.................((((((((((((((.(((((...((((....))))..))))).)))))).)))))...))).................((.......))..)))). (-22.69 = -22.38 + -0.31)


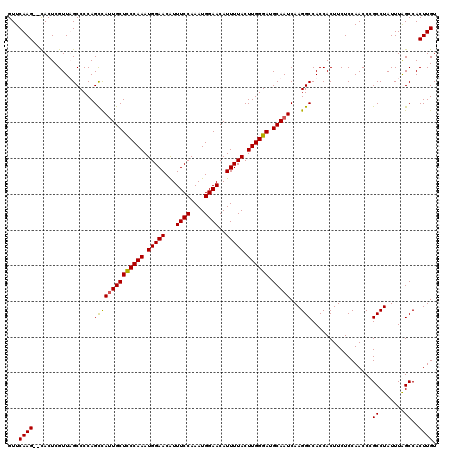
| Location | 13,147,230 – 13,147,348 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -33.40 |
| Energy contribution | -34.03 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147230 118 - 23771897 ACAAGUGGCUAAAUAGGCAGGUUGGAGAAGUGGUGGCCUUGAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCUAACGAGUA--CUUGAAC .(((((((((.....)))............((.(((((((.(((((.((((((((..((((((((....)).))))))..))))))))))))).)...)))))).))..))--))))... ( -36.70) >DroSec_CAF1 134884 120 - 1 ACAAGUGGCUAAAUAGGCGGGUUGGAGAAGUGGUGGCCUUGAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCAAACGAGUGCGCUUGAAC .(((((((((.....)))...........((..((.((((.(((((.((((((((..((((((((....)).))))))..))))))))))))).)..))).)).)).....))))))... ( -37.90) >DroSim_CAF1 134757 118 - 1 ACAAGUGGCUAAAUAGGCGGGUUGGAGAAGUGGUGGCCUUGAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGAAGCAAUGACUGGGGCAAACGAGUG--CUUGAAC .(((.(.(((.....))).).))).....((...(((((((.((((..((((.((....(((((((..((((.....))))..))).))))...)))))))))).)))).)--))...)) ( -37.00) >DroYak_CAF1 136443 118 - 1 ACAAGUGGCCAAAUAGGCGGGUUGGAGAAGUGGUGGCCUUGAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAACGUUCCAUUUGGGAGCAUUAGCUGGGGCUAACGAGUG--CUUGAAC .(((.(.(((.....))).).))).....((...(((((((..(((.((((((((..((((((((....)).))))))..)))))))))))(((((....))))))))).)--))...)) ( -41.30) >consensus ACAAGUGGCUAAAUAGGCGGGUUGGAGAAGUGGUGGCCUUGAUUGCAUCCCAAGUAAAAUGUUCCAUUUGGAAAUGUUCCAUUUGGGAGCAAUGGCUGGGGCAAACGAGUG__CUUGAAC .(((.(.(((.....))).).))).........(((((((.(((((.((((((((..((((((((....)).))))))..)))))))))))))....))))))).((((....))))... (-33.40 = -34.03 + 0.62)


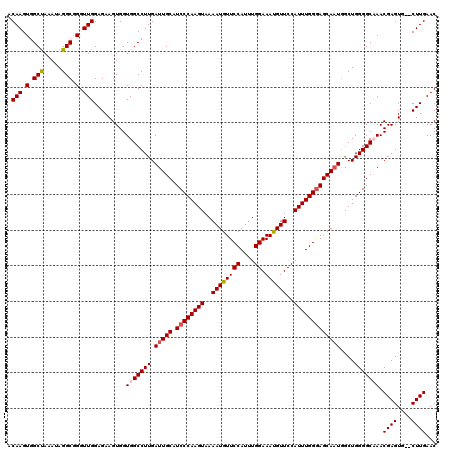
| Location | 13,147,348 – 13,147,465 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -36.35 |
| Energy contribution | -36.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13147348 117 - 23771897 GACGACGAGACGGGGCGUGGUCGGGUCUCAGGAUGCUCGACGCACGCAUUAAAACAAUUUAAAAUUCCCAAAGACCGAAACACUAGGC---AACCCACUACGGUGGGUGGCAUUGCCCAG ...........(((((((((((((((........))))))).))))).(((((....)))))....))).............((.(((---(((((((....)))))).....)))).)) ( -40.60) >DroSec_CAF1 135004 117 - 1 GACGACGAGACGGGGCGUGGUCGGGUCUCAGGAUGCUCGACGCACGCAUUAAAACAAUUUAAAAUUCCCAAAGACCGAAACACUAGGC---AACCCACAACGGUGGGUGGCAUCGCCCAG .....(((...(((((((((((((((........))))))).))))).(((((....)))))....))).................((---.((((((....)))))).)).)))..... ( -38.10) >DroSim_CAF1 134875 117 - 1 GACGACGAGACGGGGCGUGGUCGGGUCUCAGGAUGCUCGACGCACGCAUUAAAACAAUUUAAAAUUCCCAAAGACCGAAACACUAGGC---AACCCACAACGGUGGGUGGCAUCGCCCAG .....(((...(((((((((((((((........))))))).))))).(((((....)))))....))).................((---.((((((....)))))).)).)))..... ( -38.10) >DroYak_CAF1 136561 120 - 1 GACGACGAGACGGGGCGUGGUCGGGUCUCAGGAUGCUCGACGCACGCAUUAAAACAAUUUAAAAUUCCCAAAGACCGAAACACUAGACGACGACCCACUACGGUGGGUGGCAUCGCACAG ...(.(((...(((((((((((((((........))))))).))))).(((((....)))))....)))...................(.(.((((((....)))))).)).)))).... ( -40.30) >consensus GACGACGAGACGGGGCGUGGUCGGGUCUCAGGAUGCUCGACGCACGCAUUAAAACAAUUUAAAAUUCCCAAAGACCGAAACACUAGGC___AACCCACAACGGUGGGUGGCAUCGCCCAG ...........(((((((((((((((........))))))).))))).(((((....)))))....))).............((.(((....((((((....))))))......))).)) (-36.35 = -36.85 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:32 2006