| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,143,608 – 13,143,727 |
| Length | 119 |
| Max. P | 0.933897 |

| Location | 13,143,608 – 13,143,727 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143608 119 + 23771897 CAGUGCGAGACAUUAGUGUAAGUGC-AAGGGGAACUUAAAGCAAACUUGUACAAACAAUUCAUUACUAAAAAAAUAUCUGUUAGUUUUAAUACCAUACUUUGCUCUAUGAUACGCACUGU (((((((..(((....))).((.((-((((((...(((((((.(((..(((.......................)))..))).)))))))..))...)))))).))......))))))). ( -24.50) >DroSec_CAF1 131304 107 + 1 CAGUGCGAGACAUUAGUGUAAGUGC-AAGGGGUAAUUUAAGCAAAC---------CAUUUCAUUACUGA---AAUAUCUGCUAGUUUACAUACCAUAUUUUGCGCUAUGAUACGCACUGU (((((((...(((.((((((((...-..((.((((....((((...---------.((((((....)))---)))...))))...))))...))....)))))))))))...))))))). ( -31.10) >DroSim_CAF1 131205 108 + 1 CAGUGUGAGACAUAAGUGUAAGUGCGAAAGGGAACUUUAAGCAAAU---------CAUUUCAAUAGUGA---AAUAUCUGCUAGUUUAAAUACCAUUUUUUGCGCUAUGAUACGCACUGU (((((((...((((.(((((((..(....).(((((...((((.((---------.((((((....)))---))))).)))))))))...........)))))))))))...))))))). ( -29.00) >DroEre_CAF1 128159 113 + 1 CAGUGCGAGACAUUAGUGUAAGUAC-A---GAAACUUUGAGCAAACUUAUUCAAACAGUUCAUUACUCA---GAUAUCUGUUAGCUUAAAUACCAAAUUUUUCGCUAUGAUACGCACUGU (((((((...(((.(((((((((((-(---((...((((((..((((.........)))).....))))---))..)))))..)))))..............)))))))...))))))). ( -24.93) >DroYak_CAF1 132656 115 + 1 CAGUGCGAGACAUCAGUGCAAGUAC-AAGGGGAACUUU-AGCAAACUUAUUCAAACAACCCAUUACUUA---AAUAUCUGUUAGCUUAAAUACUUAAUUUUUAGUUAUGAUACGCACUGU (((((((............(((((.-..(((.......-...................)))..))))).---..((((...(((((.(((........))).))))).))))))))))). ( -22.47) >consensus CAGUGCGAGACAUUAGUGUAAGUGC_AAGGGGAACUUUAAGCAAACUU_U_CAAACAAUUCAUUACUGA___AAUAUCUGUUAGUUUAAAUACCAUAUUUUGCGCUAUGAUACGCACUGU (((((((...(((.((((((((.......((....(((((((.(((.................................))).)))))))..))....)))))))))))...))))))). (-17.09 = -17.61 + 0.52)

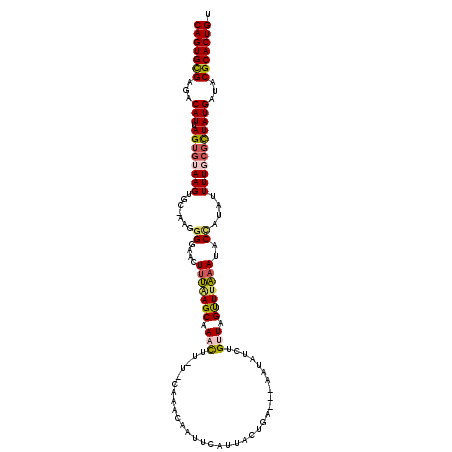
| Location | 13,143,608 – 13,143,727 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143608 119 - 23771897 ACAGUGCGUAUCAUAGAGCAAAGUAUGGUAUUAAAACUAACAGAUAUUUUUUUAGUAAUGAAUUGUUUGUACAAGUUUGCUUUAAGUUCCCCUU-GCACUUACACUAAUGUCUCGCACUG .(((((((...(((.(((((((.(.((((......))))(((((((....((((....)))).)))))))...).)))))))(((((.(.....-).))))).....)))...))))))) ( -28.00) >DroSec_CAF1 131304 107 - 1 ACAGUGCGUAUCAUAGCGCAAAAUAUGGUAUGUAAACUAGCAGAUAUU---UCAGUAAUGAAAUG---------GUUUGCUUAAAUUACCCCUU-GCACUUACACUAAUGUCUCGCACUG .(((((((...(((((.((((.....((((........((((((((((---(((....)))))).---------))))))).....))))..))-)).)).......)))...))))))) ( -31.33) >DroSim_CAF1 131205 108 - 1 ACAGUGCGUAUCAUAGCGCAAAAAAUGGUAUUUAAACUAGCAGAUAUU---UCACUAUUGAAAUG---------AUUUGCUUAAAGUUCCCUUUCGCACUUACACUUAUGUCUCACACUG .(((((.(...(((((((....(((.((.((((.....((((((((((---(((....)))))).---------)))))))..)))).)).)))))).........))))...).))))) ( -21.20) >DroEre_CAF1 128159 113 - 1 ACAGUGCGUAUCAUAGCGAAAAAUUUGGUAUUUAAGCUAACAGAUAUC---UGAGUAAUGAACUGUUUGAAUAAGUUUGCUCAAAGUUUC---U-GUACUUACACUAAUGUCUCGCACUG .(((((((...((((((...((((.....))))..))))(((((..(.---(((((...(((((.........)))))))))).)...))---)-))...........))...))))))) ( -27.80) >DroYak_CAF1 132656 115 - 1 ACAGUGCGUAUCAUAACUAAAAAUUAAGUAUUUAAGCUAACAGAUAUU---UAAGUAAUGGGUUGUUUGAAUAAGUUUGCU-AAAGUUCCCCUU-GUACUUGCACUGAUGUCUCGCACUG .(((((((...(((((((.....((((((((((........)))))))---)))......))))((..(.(((((...((.-...))....)))-)).)..))....)))...))))))) ( -24.90) >consensus ACAGUGCGUAUCAUAGCGCAAAAUAUGGUAUUUAAACUAACAGAUAUU___UCAGUAAUGAAAUGUUUG_A_AAGUUUGCUUAAAGUUCCCCUU_GCACUUACACUAAUGUCUCGCACUG .(((((((...(((...(((((...((((......))))............(((....)))..............)))))...((((.(......).))))......)))...))))))) (-12.16 = -12.96 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:26 2006