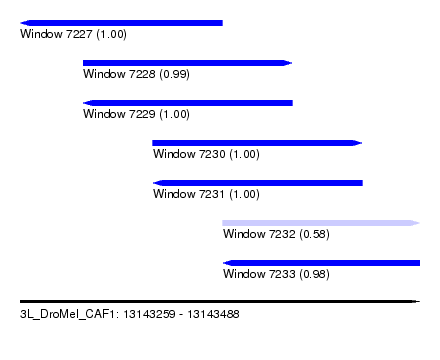
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,143,259 – 13,143,488 |
| Length | 229 |
| Max. P | 0.999912 |

| Location | 13,143,259 – 13,143,375 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143259 116 - 23771897 UGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCGCAGGGAGGAGGA----AACCGAUAAAUCUGUGGCAGAGCA ..(((((((((((((((....))))))............................(((((.....((((......))))..........(..----..)))))).....).)))))))). ( -27.70) >DroSec_CAF1 130955 116 - 1 UGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCGCUGGGAGGAGGA----AACCGAUAAAUCUGUGGCAGAGCA ..(((((((((((((((....))))))............................(((((.....((((......))))..........(..----..)))))).....).)))))))). ( -27.70) >DroSim_CAF1 130856 116 - 1 UGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCGUUGGGAGGAGGA----AACCGAUAAAUCUGUGGCAGAGCA ..(((((((((((((((....))))))............................(((((..(((((((......))))))).......(..----..)))))).....).)))))))). ( -30.20) >DroEre_CAF1 127810 116 - 1 UGGUUUUGCCCACAAACGAAAGUUUGUUAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAGAGGUCGCAGGGAGGAGGA----AACCGAUAAAUCUGCGGCAGAGCA ..((((((((.((((((....))))))((((((((...........................))))))))(((((.((((.........(..----..)))))...))))))))))))). ( -31.13) >DroYak_CAF1 132296 120 - 1 UGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAGAGUUCGCUGGCAGGAGGAGGAGGACCGAUAAAUCUGCGGCAGAGCA ....((((((.((((((....)))))).(((((((.........)))))))..................))))))((((((((.((((....((....))......)))))))).)))). ( -29.90) >consensus UGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCGCUGGGAGGAGGA____AACCGAUAAAUCUGUGGCAGAGCA ..(((((((((((((((....)))))).(((((((...............((......((((..((.....))...)))).......))((.......)))))))))..).)))))))). (-24.82 = -25.02 + 0.20)


| Location | 13,143,295 – 13,143,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143295 120 + 23771897 CGAACUAUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCUACCUU ((((..(((.....))).))))......((((((((.........))))))))((((((....))))))(((((((......)))((((((.........)))))))))).......... ( -25.10) >DroSec_CAF1 130991 120 + 1 CGAACUAUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCUAAGCUAUGGCCACGCCCUCGCCACCUU ((((..(((.....))).))))......((((((((.........))))))))((((((....))))))(((((((......)))((((((.........)))))))))).......... ( -25.10) >DroSim_CAF1 130892 120 + 1 CGAACUAUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUU ((((..(((.....))).))))......((((((((.........))))))))((((((....))))))(((((((......)))((((((.........)))))))))).......... ( -25.10) >DroEre_CAF1 127846 120 + 1 CGACCUCUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUAACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUU (((....((((..................(((((((.........))))))).((((((....)))))).))))...........((((((.........))))))....)))....... ( -23.20) >DroYak_CAF1 132336 120 + 1 CGAACUCUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUU (((....((((.................((((((((.........))))))))((((((....)))))).))))...........((((((.........))))))....)))....... ( -24.50) >consensus CGAACUAUGCCAUUCAUUUUCGAUAUUUGAUUUAUUUUAAUUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUU (((....((((.................((((((((.........))))))))((((((....)))))).))))...........((((((.........))))))....)))....... (-23.94 = -24.14 + 0.20)

| Location | 13,143,295 – 13,143,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -30.94 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143295 120 - 23771897 AAGGUAGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCG ..((((..(((((((..((((((..((((.....))))))))))..)))))((((((....)))))).......................))...))))......((((......)))). ( -32.60) >DroSec_CAF1 130991 120 - 1 AAGGUGGCGAGGGCGUGGCCAUAGCUUAGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCG ..((((..((((((((((((.........))))))(((......)))))))((((((....)))))).......................))...))))......((((......)))). ( -30.80) >DroSim_CAF1 130892 120 - 1 AAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCG ..((((..(((((((..((((((..((((.....))))))))))..)))))((((((....)))))).......................))...))))......((((......)))). ( -32.30) >DroEre_CAF1 127846 120 - 1 AAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUUAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAGAGGUCG ..((((..(((((((..((((((..((((.....))))))))))..)))))((((((....)))))).......................))...))))..........(((....))). ( -31.40) >DroYak_CAF1 132336 120 - 1 AAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAGAGUUCG ..((((..(((((((..((((((..((((.....))))))))))..)))))((((((....)))))).......................))...))))......((((......)))). ( -32.30) >consensus AAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAAUUAAAAUAAAUCAAAUAUCGAAAAUGAAUGGCAUAGUUCG ..((((..(((((((..((((((..((((.....))))))))))..)))))((((((....)))))).......................))...))))......((((......)))). (-30.94 = -31.18 + 0.24)


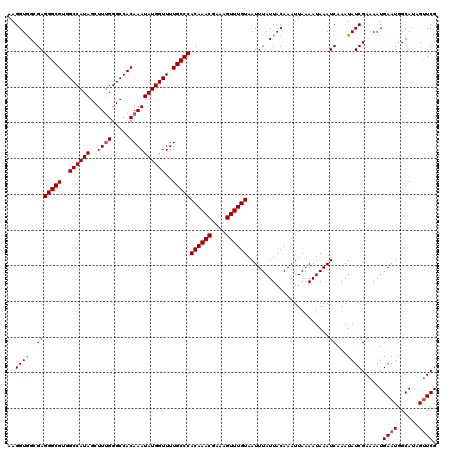
| Location | 13,143,335 – 13,143,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143335 120 + 23771897 UUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCUACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGC (((((((....)))))))........(((((......)))))...((((((.........))))))((((((((((((.....)))((((..((((....))))..)))))))))).))) ( -37.90) >DroSec_CAF1 131031 120 + 1 UUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCUAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCUGGCGGC .............((((((....))))))(((((((......)))((((((.........))))))))))((((((((.....))(((((..((((....))))..)))))..)))))). ( -34.50) >DroSim_CAF1 130932 120 + 1 UUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCUGGCGGC .............((((((....))))))(((((((......)))((((((.........))))))))))((((((((.....))(((((..((((....))))..)))))..)))))). ( -34.50) >DroEre_CAF1 127886 120 + 1 UUUGUAAUAAAUAACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGC ........(((((((((((....))))))((......)).)))))((((((.........))))))((((((((.(((.....)))((((..((((....))))..)))).))))).))) ( -35.50) >DroYak_CAF1 132376 120 + 1 UUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGUAGGCGGC .............((((((....))))))(((((((......)))((((((.........))))))))))((((((((.....)))((((..((((....))))..))))....))))). ( -34.30) >consensus UUUGUAAUAAAUUACAAACUUUCGUUUGUGGGCAAAACCAUAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGC .............((((((....))))))(((((((......)))((((((.........))))))))))((((((((.....)))((((..((((....))))..))))....))))). (-33.84 = -33.68 + -0.16)

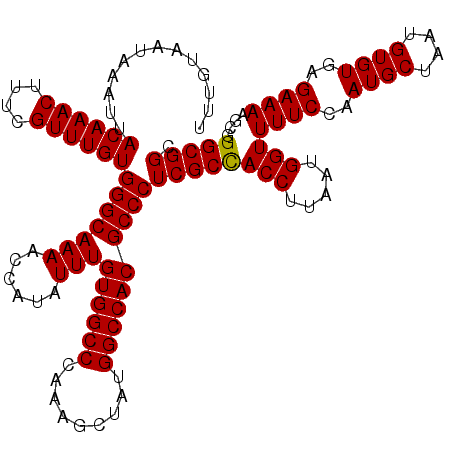

| Location | 13,143,335 – 13,143,455 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -39.88 |
| Energy contribution | -40.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143335 120 - 23771897 GCCGCCCGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUAGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAA ((..((.(((.............)))..(((....)))....))..))..(((((..((((((..((((.....))))))))))..)))))((((((....))))))............. ( -37.62) >DroSec_CAF1 131031 120 - 1 GCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUAGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAA ((((((.(((.............)))..(((....)))....))))))..((((((((((.........))))))(((......)))))))((((((....))))))............. ( -42.22) >DroSim_CAF1 130932 120 - 1 GCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAA ((((((.(((.............)))..(((....)))....))))))..(((((..((((((..((((.....))))))))))..)))))((((((....))))))............. ( -43.72) >DroEre_CAF1 127886 120 - 1 GCCGCCCGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUUAUUUAUUACAAA ((((((.(((.............)))..(((....)))....))))))..(((((..((((((..((((.....))))))))))..)))))((((((....))))))............. ( -42.32) >DroYak_CAF1 132376 120 - 1 GCCGCCUACUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAA (((((((..((((((..............))))))......)))))))..(((((..((((((..((((.....))))))))))..)))))((((((....))))))............. ( -40.54) >consensus GCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUAUGGUUUUGCCCACAAACGAAAGUUUGUAAUUUAUUACAAA ((((((.(((.............)))..(((....)))....))))))..(((((..((((((..((((.....))))))))))..)))))((((((....))))))............. (-39.88 = -40.48 + 0.60)

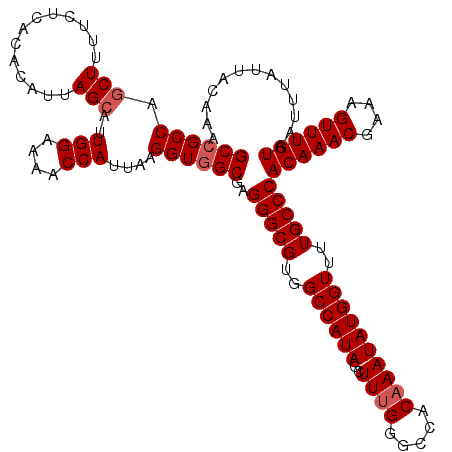

| Location | 13,143,375 – 13,143,488 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -29.01 |
| Energy contribution | -29.21 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143375 113 + 23771897 UAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCUACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGCAAAAAGCGAAAGAAAAGUGAGAGUCU-------CUCGGCA .....((((((.........))))))((((((((((((.....)))((((..((((....))))..)))))))))).)))......(....)....(((((.....-------))).)). ( -35.40) >DroSec_CAF1 131071 113 + 1 UAUUUGUGGCCCUAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCUGGCGGCAAAAAGCGAAAGAAAAGUGAGAGUCU-------AUCGGCA .....((((((.........))))))(((((((((.((.....(.(((((..((((....))))..))))).).)).)))......(....)........)))...-------...))). ( -32.40) >DroSim_CAF1 130972 113 + 1 UAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCUGGCGGCAAAAAGCGAAAGAAAAGUGAGAGUCU-------AUCGGCA .....((((((.........))))))(((((((((.((.....(.(((((..((((....))))..))))).).)).)))......(....)........)))...-------...))). ( -32.40) >DroEre_CAF1 127926 113 + 1 UAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGCAAAAGGCGAAAGAAAAGUGAGAGUCU-------CUCCGCA .....((((((.........))))))((((((((.(((.....)))((((..((((....))))..)))).))))).)))......(....)....(((.(((...-------)))))). ( -35.00) >DroYak_CAF1 132416 120 + 1 UAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGUAGGCGGCAAAAAGCGAACGAAAAGUGAGAGUCUCUCGUCUCUCGGCA .....((((((.........))))))(((...(((.(((....(.(((((..((((....))))..))))).)))).)))..................(((((.(....).)))))))). ( -37.30) >consensus UAUUUGUGGCCCAAAGCUAUGGCCACGCCCUCGCCACCUUAAUGGUUUUCCAAUGCUAAUGUGUGAGAAAAGCGGGCGGCAAAAAGCGAAAGAAAAGUGAGAGUCU_______CUCGGCA .....((((((.........))))))(((((((((.(((....(.(((((..((((....))))..))))).)))).)))......(....)........))).............))). (-29.01 = -29.21 + 0.20)



| Location | 13,143,375 – 13,143,488 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13143375 113 - 23771897 UGCCGAG-------AGACUCUCACUUUUCUUUCGCUUUUUGCCGCCCGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUAGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUA .((((((-------(((..........))))))....(((((..((.(((.............)))..(((....)))....))..))))))))((((((.........))))))..... ( -32.82) >DroSec_CAF1 131071 113 - 1 UGCCGAU-------AGACUCUCACUUUUCUUUCGCUUUUUGCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUAGGGCCACAAAUA .(((((.-------(((..........))).))....(((((((((.(((.............)))..(((....)))....))))))))))))((((((.........))))))..... ( -35.52) >DroSim_CAF1 130972 113 - 1 UGCCGAU-------AGACUCUCACUUUUCUUUCGCUUUUUGCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUA .(((((.-------(((..........))).))....(((((((((.(((.............)))..(((....)))....))))))))))))((((((.........))))))..... ( -35.52) >DroEre_CAF1 127926 113 - 1 UGCGGAG-------AGACUCUCACUUUUCUUUCGCCUUUUGCCGCCCGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUA ....(((-------(((..........))))))((((((.((((((.(((.............)))..(((....)))....))))))))))))((((((.........))))))..... ( -38.82) >DroYak_CAF1 132416 120 - 1 UGCCGAGAGACGAGAGACUCUCACUUUUCGUUCGCUUUUUGCCGCCUACUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUA .((((((((.(....).)))))...............((((((((((..((((((..............))))))......)))))))))))))((((((.........))))))..... ( -40.04) >consensus UGCCGAG_______AGACUCUCACUUUUCUUUCGCUUUUUGCCGCCAGCUUUUCUCACACAUUAGCAUUGGAAAACCAUUAAGGUGGCGAGGGCGUGGCCAUAGCUUUGGGCCACAAAUA .................................((((((.((((((.(((.............)))..(((....)))....))))))))))))((((((.........))))))..... (-30.42 = -30.66 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:24 2006