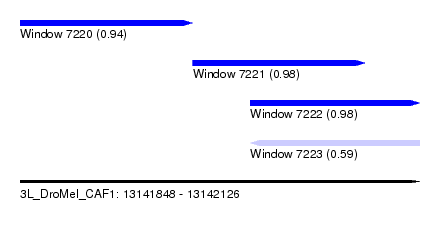
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,141,848 – 13,142,126 |
| Length | 278 |
| Max. P | 0.983416 |

| Location | 13,141,848 – 13,141,968 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13141848 120 + 23771897 AGGCACAAAAAGGCAACAAAGGUGCAUAUCUCCUCCUUUCGGACUUUCGGACACAACAAAGCCGGUGUUCUGGCAAAAAGGAAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC ..((((.....(....)....))))........(((....)))(((((((..........(((((....))))).............(((((....))))).....)).)))))...... ( -25.10) >DroSec_CAF1 129521 120 + 1 AGGCACAAAAAGGAAACAACGGUGCAUAUCUCCUCCUUUCGGACUUACGGACACAACAAAGCCGGUGUUCUGGCAAAAAGGGAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC .((........(....)...(((.(((((.((((...((((......)))).........(((((....))))).....)))).)))))....)))..........))............ ( -26.30) >DroSim_CAF1 129439 120 + 1 AGGCACAAAAAGGCAACAAAGGUGCAUAUCUCCUCCUUUCGGACUUUCGGACACAACAAAGCCGGUGUUCUGGCAAAAAGGGAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC ..((((.....(....)....))))........(((....)))(((((............(((((....))))).....((......(((((....))))).....)).)))))...... ( -25.40) >DroEre_CAF1 126424 119 + 1 AGGCACAAAAAGGCAACAAAGGUGCAUAUCUCCUCCUUUCGGACUUUCGGACACAACAAAGCCGGUGUUGUGGCAAAAAG-AAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC ..((((.....(....)....))))..........(((((((.((((..(.(((((((.......))))))).)..))))-......(((((....))))).....)).)))))...... ( -25.80) >DroYak_CAF1 130884 119 + 1 AGGCACAAAAAGGCAACAAAGGUGCAUAUCUCCUCCUUUCGGACGUUCGGACACAACAAAGCCGGUGUUGUGGCAAAAAG-AAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC ..((((.....(....)....))))..........(((((((.........(((((((.......)))))))........-......(((((....))))).....)).)))))...... ( -23.10) >consensus AGGCACAAAAAGGCAACAAAGGUGCAUAUCUCCUCCUUUCGGACUUUCGGACACAACAAAGCCGGUGUUCUGGCAAAAAGGAAAAUAUGAAUAACCAUUCAAUCAACCAGAAAGAACAAC ..((((.....(....)....))))........(((....)))(((((((..........(((((....))))).............(((((....))))).....)).)))))...... (-21.70 = -22.50 + 0.80)

| Location | 13,141,968 – 13,142,088 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.25 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13141968 120 + 23771897 AAUUUCAACGAGGUGGAGCAAAAAAUAUGGCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAA ........((.((.((.(((.......(((....)))((((...(((((.....).((((((((..(((....))).))))))))..))))..))))......))).))))))....... ( -24.40) >DroSec_CAF1 129641 120 + 1 AAUUUCAACGAGUGGGAGCAAAAAAUAUGGCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAA ........((.(.(((.(((.......(((....)))((((...(((((.....).((((((((..(((....))).))))))))..))))..))))......))).))))))....... ( -24.40) >DroSim_CAF1 129559 120 + 1 AAUUUCAACGAGGGGGAGCAAAAAAUAUGGCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGGUAUAAA ...........(((((.(((.......(((....)))((((...(((((.....).((((((((..(((....))).))))))))..))))..))))......))).)))))........ ( -29.90) >DroEre_CAF1 126543 119 + 1 AAUUUCAACGAGG-GGAGCAAAAAAUAUGUCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAA ........((.((-((.(((........(((((.((.........)))))))....((((((((..(((....))).))))))))..................))).))))))....... ( -29.70) >DroYak_CAF1 131003 120 + 1 AAUUUCAACGAGGGGGAGCAAAAAAUAUGUCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAA ...........(((((.(((........(((((.((.........)))))))....((((((((..(((....))).))))))))..................))).)))))........ ( -31.40) >consensus AAUUUCAACGAGGGGGAGCAAAAAAUAUGGCCAGUCAUUAAUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAA ...........(((((.(((...............((((......))))((((...((((((((..(((....))).)))))))).)))).............))).)))))........ (-25.24 = -25.84 + 0.60)



| Location | 13,142,008 – 13,142,126 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -26.87 |
| Energy contribution | -26.71 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13142008 118 + 23771897 AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAAGCGGCGAACCAUAUUUUUUGCCAUGC--UUAAGCCGCACA ((((((..((......((((((((..(((....))).))))))))....(((............)))))..))))))...((((((((........))))))..((--....)).))... ( -27.70) >DroSec_CAF1 129681 120 + 1 AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAAGCGGCGAACCAUAUUUUUUACCAUGCUCUUAAGCCGCACA ......((((......((((((((..(((....))).)))))))).........(((((((((((((((.............))))...)))))))))))))))(((....)))...... ( -27.52) >DroSim_CAF1 129599 120 + 1 AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGGUAUAAAGCGGCGAACCAUAUUUUUUACCAUGCUCUUAAGCCGCACA ......((((......((((((((..(((....))).)))))))).........(((((((((((((((.............))))...)))))))))))))))(((....)))...... ( -27.52) >DroEre_CAF1 126582 118 + 1 AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAAGCGGCGAACCAUAUUUUUUACCAUGC--UUAAGCCGCACA ((((((..((......((((((((..(((....))).))))))))....(((............)))))..))))))...(((((....(((..........))).--....)))))... ( -27.40) >DroYak_CAF1 131043 118 + 1 AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAAGCGGCGAACCAUAUUUUUUACCAACC--UUAAGCCGCACA ((((((..((......((((((((..(((....))).))))))))....(((............)))))..))))))...(((((.....................--....)))))... ( -26.91) >consensus AUAUCGAUGGAUUACAUGCGUUCAGCAUUUUACGAUAUGAGCGCAAAAUCGUUUUAAAAAAUAUGCGCCCCCGAUAUAAAGCGGCGAACCAUAUUUUUUACCAUGC__UUAAGCCGCACA ((((((..((......((((((((..(((....))).))))))))....(((............)))))..))))))...(((((...........................)))))... (-26.87 = -26.71 + -0.16)


| Location | 13,142,008 – 13,142,126 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13142008 118 - 23771897 UGUGCGGCUUAA--GCAUGGCAAAAAAUAUGGUUCGCCGCUUUAUAUCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU ...(((((..((--.((((........)))).)).)))))...((((((((((((((..................)))))))..........((((.....))))........))))))) ( -31.07) >DroSec_CAF1 129681 120 - 1 UGUGCGGCUUAAGAGCAUGGUAAAAAAUAUGGUUCGCCGCUUUAUAUCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU .((((((((....))).((.(((((((((((((((.(((........))))))).)))))))))))...))....)))))..((((((....((((.....)))).........)))))) ( -30.52) >DroSim_CAF1 129599 120 - 1 UGUGCGGCUUAAGAGCAUGGUAAAAAAUAUGGUUCGCCGCUUUAUACCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU .((((((((....))).((.(((((((((((((((.(((........))))))).)))))))))))...))....)))))..((((((....((((.....)))).........)))))) ( -30.72) >DroEre_CAF1 126582 118 - 1 UGUGCGGCUUAA--GCAUGGUAAAAAAUAUGGUUCGCCGCUUUAUAUCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU .(((((.....(--((.((.(((((((((((((((.(((........))))))).)))))))))))...))((((((((.......)))))))))))...)))))............... ( -31.70) >DroYak_CAF1 131043 118 - 1 UGUGCGGCUUAA--GGUUGGUAAAAAAUAUGGUUCGCCGCUUUAUAUCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU .....(((.(((--(((((.(((((((((((((((.(((........))))))).)))))))))))...)))))))).))).((((((....((((.....)))).........)))))) ( -32.02) >consensus UGUGCGGCUUAA__GCAUGGUAAAAAAUAUGGUUCGCCGCUUUAUAUCGGGGGCGCAUAUUUUUUAAAACGAUUUUGCGCUCAUAUCGUAAAAUGCUGAACGCAUGUAAUCCAUCGAUAU .(((((((......))..(((((((((((((((((.(((........))))))).))))))))))......((((((((.......)))))))))))...)))))............... (-27.46 = -27.50 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:14 2006