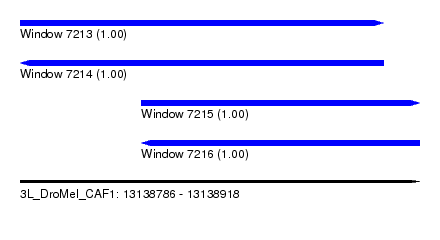
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,138,786 – 13,138,918 |
| Length | 132 |
| Max. P | 0.999920 |

| Location | 13,138,786 – 13,138,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13138786 120 + 23771897 CACCCAGCGAUUGGCGCCACACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGG ...(((((((..((((...(.(((.(((.........(((((((....)))))))((.....)).........))).))))...))))((((((.....))))))........))))))) ( -30.90) >DroSec_CAF1 126529 120 + 1 CACCCAGCGAUUGGCGCCAUACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGUUUGUGCAACAACUGCACAUUAUUUAUUCGCUGG ...(((((((.(((.(((...(((.(((.........(((((((....)))))))((.....)).........))).)))))).....((((((.....))))))....))).))))))) ( -30.30) >DroSim_CAF1 126484 120 + 1 CACCCAGCGAUUGGCGCCACACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGG ...(((((((..((((...(.(((.(((.........(((((((....)))))))((.....)).........))).))))...))))((((((.....))))))........))))))) ( -30.90) >DroEre_CAF1 123415 120 + 1 CACCCAGCGAUUGGCGCCACACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACCCUCGUGCAACAACUGCACAUUAUUUAUUCGCUGG ...(((((((..((.(((...(((.(((.........(((((((....)))))))((.....)).........))).)))))).))...(((((.....))))).........))))))) ( -32.20) >DroYak_CAF1 127769 120 + 1 CACCCAGCGAUUGGCGCCACACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUCGUGCAACAACUGCAUAUUAUUUAUUCGCUGG ...(((((((...(((.........)))................(((((((((((((.............(((.....)))((((....)))).......)))))))))))))))))))) ( -30.90) >consensus CACCCAGCGAUUGGCGCCACACAUAUGCAACUAAUUGAUAUUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGG ...(((((((.(((.(((...(((.(((.........(((((((....)))))))((.....)).........))).))))))......(((((.....))))).....))).))))))) (-29.42 = -29.26 + -0.16)

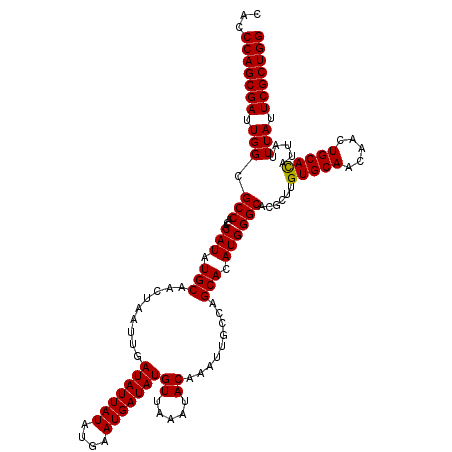
| Location | 13,138,786 – 13,138,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -37.01 |
| Energy contribution | -36.97 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13138786 120 - 23771897 CCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUGUGGCGCCAAUCGCUGGGUG (((((((........(((((((...))))))).(.((((((((((((.(((((((.(((((.................))))).)))).))))))))))...))))))..)))))))... ( -38.33) >DroSec_CAF1 126529 120 - 1 CCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAACGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUAUGGCGCCAAUCGCUGGGUG (((((((........(((((((...)))))))...((((((((((((.(((((((.(((((.................))))).)))).))))))))))...)))))...)))))))... ( -38.03) >DroSim_CAF1 126484 120 - 1 CCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUGUGGCGCCAAUCGCUGGGUG (((((((........(((((((...))))))).(.((((((((((((.(((((((.(((((.................))))).)))).))))))))))...))))))..)))))))... ( -38.33) >DroEre_CAF1 123415 120 - 1 CCAGCGAAUAAAUAAUGUGCAGUUGUUGCACGAGGGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUGUGGCGCCAAUCGCUGGGUG (((((((........(((((((...)))))))..(((((((((((((.(((((((.(((((.................))))).)))).))))))))))...))))))..)))))))... ( -41.23) >DroYak_CAF1 127769 120 - 1 CCAGCGAAUAAAUAAUAUGCAGUUGUUGCACGAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUGUGGCGCCAAUCGCUGGGUG (((((((...........(((..((..((((....)))).))..)))((((((((.(((((.................))))).)))).((..((....))..)))))).)))))))... ( -33.83) >consensus CCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAAUAUCAAUUAGUUGCAUAUGUGUGGCGCCAAUCGCUGGGUG (((((((........(((((((...)))))))...((((((((((((.(((((((.(((((.................))))).)))).))))))))))...)))))...)))))))... (-37.01 = -36.97 + -0.04)

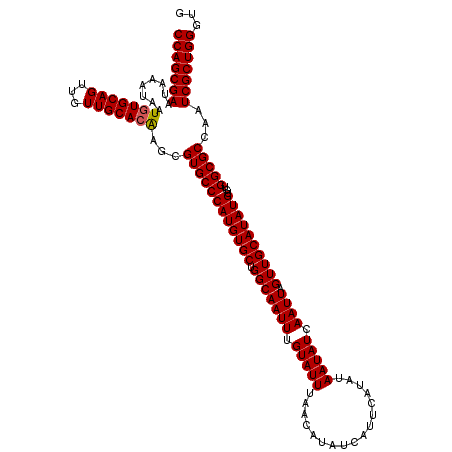

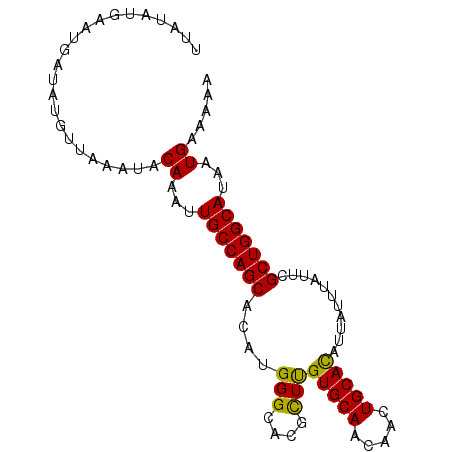
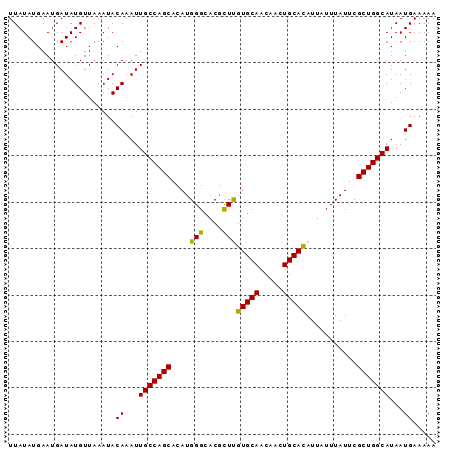
| Location | 13,138,826 – 13,138,918 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -21.30 |
| Energy contribution | -20.74 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13138826 92 + 23771897 UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((....(((....)))(((((.....)))))...........)))))))...))..... ( -21.00) >DroSec_CAF1 126569 92 + 1 UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGUUUGUGCAACAACUGCACAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((..(((....))).((((((.....))))))..........)))))))...))..... ( -21.40) >DroSim_CAF1 126524 92 + 1 UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((....(((....)))(((((.....)))))...........)))))))...))..... ( -21.00) >DroEre_CAF1 123455 92 + 1 UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACCCUCGUGCAACAACUGCACAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((....((....))..(((((.....)))))...........)))))))...))..... ( -23.30) >DroYak_CAF1 127809 92 + 1 UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUCGUGCAACAACUGCAUAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((....(((....)))(((((.....)))))...........)))))))...))..... ( -21.30) >consensus UUAUAUGAAUGAUAUGUUAAAUACAAAUUGCCAGCACAUGGGCACGCUUGUGCAACAACUGCACAUUAUUUAUUCGCUGGCAUAAUGAAAAA .......................((...(((((((....(((....)))(((((.....)))))...........)))))))...))..... (-21.30 = -20.74 + -0.56)

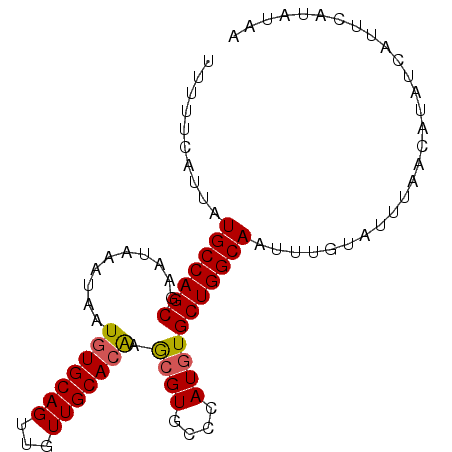
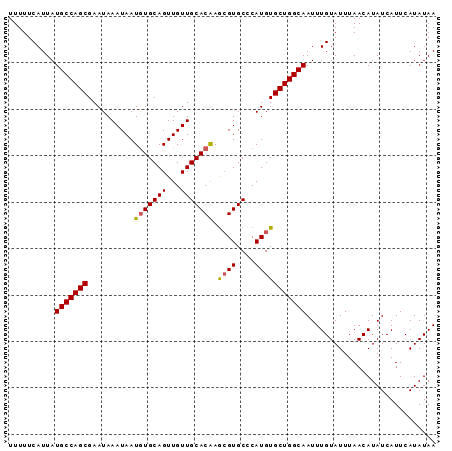
| Location | 13,138,826 – 13,138,918 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13138826 92 - 23771897 UUUUUCAUUAUGCCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........(((((((..........(((((((...))))))).((((....)))))))))))............................ ( -22.40) >DroSec_CAF1 126569 92 - 1 UUUUUCAUUAUGCCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAACGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........(((((((..........(((((((...))))))).((((....)))))))))))............................ ( -22.10) >DroSim_CAF1 126524 92 - 1 UUUUUCAUUAUGCCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........(((((((..........(((((((...))))))).((((....)))))))))))............................ ( -22.40) >DroEre_CAF1 123455 92 - 1 UUUUUCAUUAUGCCAGCGAAUAAAUAAUGUGCAGUUGUUGCACGAGGGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........((((((((.........(((((((...))))))).((....))...))))))))............................ ( -23.00) >DroYak_CAF1 127809 92 - 1 UUUUUCAUUAUGCCAGCGAAUAAAUAAUAUGCAGUUGUUGCACGAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........((((((((....((((((.....))))))((((....)))).....))))))))............................ ( -20.20) >consensus UUUUUCAUUAUGCCAGCGAAUAAAUAAUGUGCAGUUGUUGCACAAGCGUGCCCAUGUGCUGGCAAUUUGUAUUUAACAUAUCAUUCAUAUAA ..........(((((((..........(((((((...))))))).((((....)))))))))))............................ (-20.56 = -20.56 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:06 2006