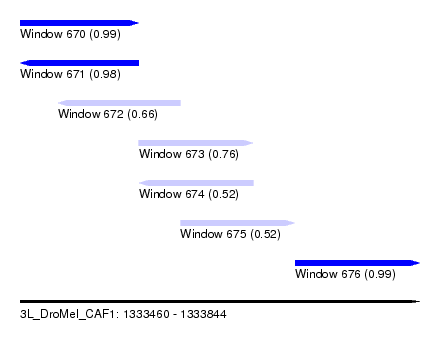
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,333,460 – 1,333,844 |
| Length | 384 |
| Max. P | 0.986266 |

| Location | 1,333,460 – 1,333,574 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -18.97 |
| Energy contribution | -20.53 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333460 114 + 23771897 CAAGCAUAAAUAUGCCAAAUACAGGCCAAU---ACACACGCACGAACUCACA--CACACACGCGUUCGAUGGAGAAUGCAAAGUAAAGUGCAGCAGGUGUGCGUGUAUGUGUGUUGGUG ...((((....)))).........((((((---(((((..((((....((((--(......((((((......))))))...((.....)).....)))))))))..))))))))))). ( -40.60) >DroSec_CAF1 5633 115 + 1 CAAGCAUAAAUAUGCCAAAUACAGGCCAGCAACACGCACGCACGAACACACA----CACACACGUUUGAUGGAGAAUGCAAAGUAAAGUGCAGCAGGUGUGCGCGUGUAUAUGUUGGUG ...((((....)))).........(((((((((((((.(((((...((((.(----(......)).)).)).....((((........))))....))))).))))))...))))))). ( -33.90) >DroEre_CAF1 5823 94 + 1 CAAGCAUAAAUAAGCCAAAUAC-------------------A------CACACACACACACACGUUUGAUGUAGAAUGCAAAGUAAAGUGCAGCAGGUGUGCGCGUGUGUGUGUUGGUG .............(((((.(((-------------------(------(((((.(.(((((((((...))))....((((........))))....))))).).)))))))))))))). ( -31.70) >consensus CAAGCAUAAAUAUGCCAAAUACAGGCCA_____AC_CACGCACGAAC_CACA__CACACACACGUUUGAUGGAGAAUGCAAAGUAAAGUGCAGCAGGUGUGCGCGUGUGUGUGUUGGUG ...((((....)))).................................(((...(((((((((((...........((((........))))(((....))))))))))))))...))) (-18.97 = -20.53 + 1.56)

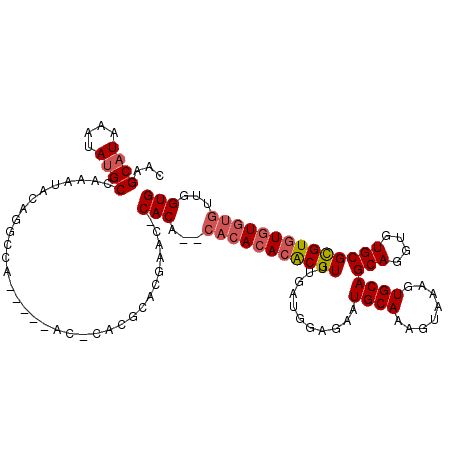
| Location | 1,333,460 – 1,333,574 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333460 114 - 23771897 CACCAACACACAUACACGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCGAACGCGUGUGUG--UGUGAGUUCGUGCGUGUGU---AUUGGCCUGUAUUUGGCAUAUUUAUGCUUG ......(((((((((((((........((((........))))(((......))).)))))))))--)))).......(((((.((---((..(((.......))))))).)))))... ( -36.40) >DroSec_CAF1 5633 115 - 1 CACCAACAUAUACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCAAACGUGUGUG----UGUGUGUUCGUGCGUGCGUGUUGCUGGCCUGUAUUUGGCAUAUUUAUGCUUG (((.((((((((((((((((....)))((((........))))...............)))))----)))))))).)))...(((((......(((.......))).....)))))... ( -34.30) >DroEre_CAF1 5823 94 - 1 CACCAACACACACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUACAUCAAACGUGUGUGUGUGUGUG------U-------------------GUAUUUGGCUUAUUUAUGCUUG .....((((((((((((((((((....((((........)))).............)))))))))))))))------)-------------------))....(((........))).. ( -33.43) >consensus CACCAACACACACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCAAACGUGUGUGUG__UGUG_GUUCGUGCGUG_GU_____UGGCCUGUAUUUGGCAUAUUUAUGCUUG (((...(((((((((..(((....)))((((........)))).............)))))))))...)))................................(((((....))))).. (-18.69 = -19.37 + 0.67)

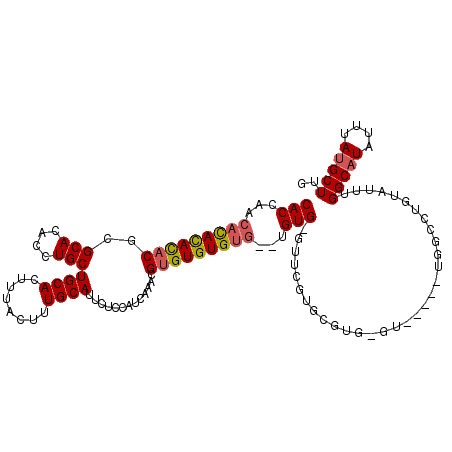
| Location | 1,333,496 – 1,333,614 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333496 118 - 23771897 AAAACAAAAACCCAUGGAAAAGAAUGUACAAACAGAAUAGCACCAACACACAUACACGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCGAACGCGUGUGUG--UGUGAGUUCGUGC ............((((((......(((....)))............(((((((((((((........((((........))))(((......))).)))))))))--))))..)))))). ( -30.50) >DroSec_CAF1 5672 116 - 1 AAAACAAAAACCACUGGAAAAGAAUGUACAAACAGAAUAGCACCAACAUAUACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCAAACGUGUGUG----UGUGUGUUCGUGC .............(((................)))....((((.((((((((((((((((....)))((((........))))...............)))))----)))))))).)))) ( -29.49) >DroEre_CAF1 5845 112 - 1 AAAACAAAAAACACUGAAAAAGAAUGUACAAACAGAAUCUCACCAACACACACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUACAUCAAACGUGUGUGUGUGUGUG------U-- ..........((((......(((.(((....)))...))).....(((((((((((((((....)))((((........)))).............)))))))))))))))------)-- ( -28.70) >consensus AAAACAAAAACCACUGGAAAAGAAUGUACAAACAGAAUAGCACCAACACACACACGCGCACACCUGCUGCACUUUACUUUGCAUUCUCCAUCAAACGUGUGUGUG__UGUG_GUUCGUGC ........................................(((...(((((((((..(((....)))((((........)))).............)))))))))...)))......... (-16.86 = -17.20 + 0.34)

| Location | 1,333,574 – 1,333,684 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333574 110 + 23771897 CUAUUCUGUUUGUACAUUCUUUUCCAUGGGUUUUUGUUUUUGGCCAAAGCCUGGCUGUUAAAGCAACAACUGCAACAACAACAGCACUGACACACAAUACACACGC--ACAC ......(((.(((.((...........(((((((.((.....)).))))))).((((((...(((.....)))......))))))..))))).)))..........--.... ( -18.90) >DroSec_CAF1 5748 101 + 1 CUAUUCUGUUUGUACAUUCUUUUCCAGUGGUUUUUGUUUUUGGCCAAAGCCUGGCUGUAUAAGCAG---------CAACAACACCGCUGGCACACAACACACACGC--ACAC ......(((.(((..........((((((((..(((((...(((....)))..(((((....))))---------)))))).))))))))..........))).))--)... ( -27.95) >DroSim_CAF1 5787 99 + 1 CUAUUCUGUUUGUACAUUCUUUUCCAGUGGUUUUUGUUUUUGGCCAAAGCCUGGCUGUUAAAGCAA---------CAACAACACCGCUGGCACACAACA--CACGA--ACAC ......(((((((..........((((((((..(((((...(((....)))..(((.....)))..---------.))))).)))))))).........--.))))--))). ( -27.15) >DroYak_CAF1 5931 109 + 1 AGAUUCUGUUUGUACAUUCUUUUUCAGUGGUUUUUGUUUUUGGCCAAAGCCUGGCUGUUAAAGCAACAC---CAACAACAACACCGCUGGCACACAACACACACACACACAC ......(((.(((..........((((((((..(((((.(((((((.....))))(((....)))....---))).))))).))))))))..........))).)))..... ( -23.05) >consensus CUAUUCUGUUUGUACAUUCUUUUCCAGUGGUUUUUGUUUUUGGCCAAAGCCUGGCUGUUAAAGCAA_________CAACAACACCGCUGGCACACAACACACACGC__ACAC ......(((.(((...........(((((((..(((((...(((....)))..(((.....)))............))))).)))))))))).)))................ (-16.19 = -16.75 + 0.56)

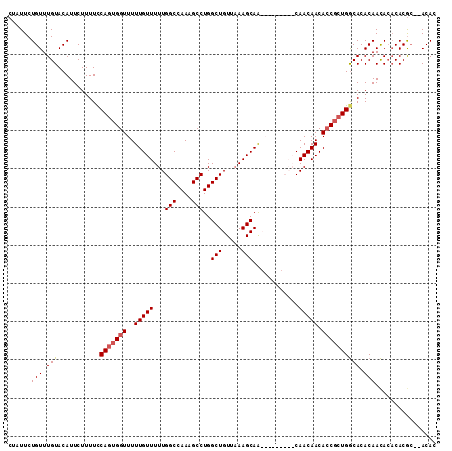
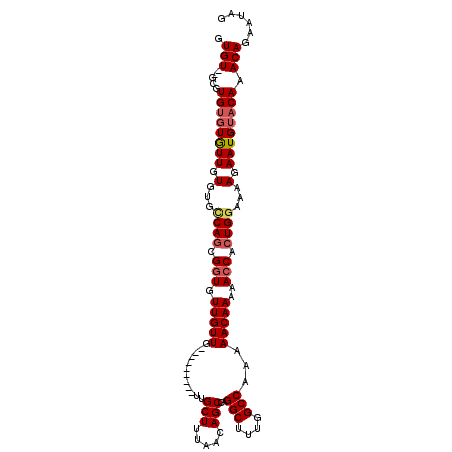
| Location | 1,333,574 – 1,333,684 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333574 110 - 23771897 GUGU--GCGUGUGUAUUGUGUGUCAGUGCUGUUGUUGUUGCAGUUGUUGCUUUAACAGCCAGGCUUUGGCCAAAAACAAAAACCCAUGGAAAAGAAUGUACAAACAGAAUAG .(((--((((.....(..((.((..(.(((((((..((.((....)).))..)))))))).(((....)))..........)).))..)......))))))).......... ( -26.60) >DroSec_CAF1 5748 101 - 1 GUGU--GCGUGUGUGUUGUGUGCCAGCGGUGUUGUUG---------CUGCUUAUACAGCCAGGCUUUGGCCAAAAACAAAAACCACUGGAAAAGAAUGUACAAACAGAAUAG .(((--...((((..((.(...((((.(((.((((((---------(((......))))..(((....)))...)))))..))).))))...).))..)))).)))...... ( -32.20) >DroSim_CAF1 5787 99 - 1 GUGU--UCGUG--UGUUGUGUGCCAGCGGUGUUGUUG---------UUGCUUUAACAGCCAGGCUUUGGCCAAAAACAAAAACCACUGGAAAAGAAUGUACAAACAGAAUAG .(((--(.(((--..((.(...((((.((((((((((---------......)))))))..(((....)))..........))).))))...).))..))).))))...... ( -30.60) >DroYak_CAF1 5931 109 - 1 GUGUGUGUGUGUGUGUUGUGUGCCAGCGGUGUUGUUGUUG---GUGUUGCUUUAACAGCCAGGCUUUGGCCAAAAACAAAAACCACUGAAAAAGAAUGUACAAACAGAAUCU ...(.(((.((((..((.(....(((.(((....((((((---((((((...))))).)).(((....)))...)))))..))).)))....).))..)))).))).).... ( -28.40) >consensus GUGU__GCGUGUGUGUUGUGUGCCAGCGGUGUUGUUG_________UUGCUUUAACAGCCAGGCUUUGGCCAAAAACAAAAACCACUGGAAAAGAAUGUACAAACAGAAUAG .(((.....((((((((.(...((((.(((.(((((............(((.....)))..(((....)))...)))))..))).))))...).)))))))).)))...... (-19.61 = -20.55 + 0.94)

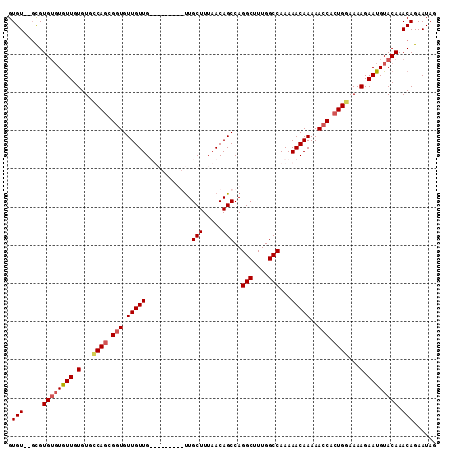
| Location | 1,333,614 – 1,333,724 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -28.55 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333614 110 + 23771897 UGGCCAAAGCCUGGCUGUUAAAGCAACAACUGCAACAACAACAGCACUGACACACAAUACACACGC--ACACACGUAAAGCUCGCCGCUGCACUGGUGUUGGUGGUGGAGGC ..(((...(((..((((((...(((.....)))......))))))((..((((.((......(((.--.....)))..(((.....)))....))))))..)))))...))) ( -28.40) >DroSec_CAF1 5788 101 + 1 UGGCCAAAGCCUGGCUGUAUAAGCAG---------CAACAACACCGCUGGCACACAACACACACGC--ACACACGUAAAGCUCGCCGCUGCACUGGUGUUAGUGGUGGAGGC ........((((.(((((....))))---------).....((((((((((((.((......(((.--.....)))..(((.....)))....)))))))))))))).)))) ( -35.40) >DroSim_CAF1 5827 99 + 1 UGGCCAAAGCCUGGCUGUUAAAGCAA---------CAACAACACCGCUGGCACACAACA--CACGA--ACACACGUAAAGCUCGCCGCUGCACUGGUGUUAGUGGUGGAGGC ........((((.(((.....)))..---------......((((((((((((.((...--.(((.--.....)))..(((.....)))....)))))))))))))).)))) ( -31.70) >DroYak_CAF1 5971 109 + 1 UGGCCAAAGCCUGGCUGUUAAAGCAACAC---CAACAACAACACCGCUGGCACACAACACACACACACACACACGUAAAGCUCGCCGAUGCACUUGUGUUGGUGGUGGAGGC ........((((((.((((.....)))))---)........((((((..(((((............((......))...((........))...)))))..)))))).)))) ( -30.10) >consensus UGGCCAAAGCCUGGCUGUUAAAGCAA_________CAACAACACCGCUGGCACACAACACACACGC__ACACACGUAAAGCUCGCCGCUGCACUGGUGUUAGUGGUGGAGGC ........((((.(((.....))).................((((((((((((.........(((........)))...((........))....)))))))))))).)))) (-28.55 = -28.42 + -0.12)

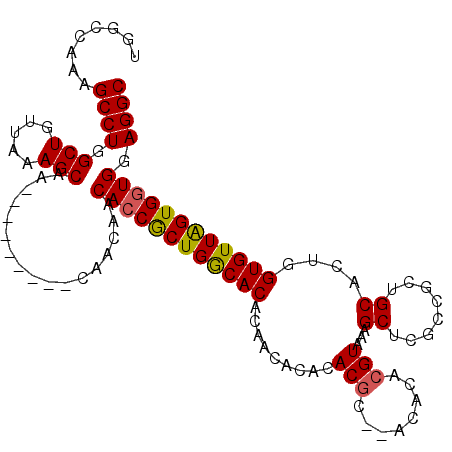
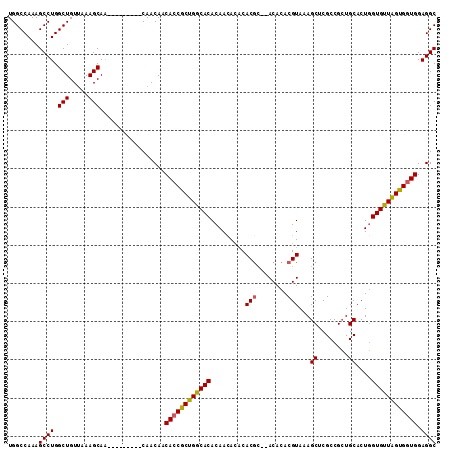
| Location | 1,333,724 – 1,333,844 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -40.58 |
| Energy contribution | -40.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1333724 120 + 23771897 AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGAGCGCCAAGGAGCUGUGGAAAGGUGUAUACUCCGCAUCCGUUGCGAAUCCUUGGCGAAUCGAACUGUGCCACA ....(((((.....)))))((((...((((......))))...((((..(((((((((((..((((..((.(....).))...))))..))...)))))))))..))))...)))).... ( -42.00) >DroSec_CAF1 5889 120 + 1 AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGAGCGCCAAGGAGCUGUGGAAAGGUGUAUACUCCGCAUCCGUUGCGAAUCCUUGGCGACUCGAAUUGUGCCACA ....(((((.....)))))((((...((((......))))...((((..(((((((((((..((((..((.(....).))...))))..))...)))))))))..))))...)))).... ( -42.10) >DroSim_CAF1 5926 120 + 1 AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGAGCGCCAAGGAGCUGUGGAAAGGUGUAUACUCCGCAUCCGUUGCGAAUCCUUGGCGACUCGAAUUGUGCCACA ....(((((.....)))))((((...((((......))))...((((..(((((((((((..((((..((.(....).))...))))..))...)))))))))..))))...)))).... ( -42.10) >DroEre_CAF1 6068 120 + 1 AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGACCGCCAAGGAGCUCUGGAAAGGUGUGUACUCCGCAUCCGUGGCGAAUCCUUGGCGACCCGAGCUAUGCCACA ....(((((.....))))).................((((...((((..((((((((((((.((((..((.(....).))...)))).)))...)))))))))..))))......)))). ( -43.30) >DroYak_CAF1 6080 120 + 1 AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGACCGCCAAGGAGCUCUGGGAAGGUGUGUACUCCGCAUCCGUGGCGAGUCCUUGGCGACUGGAGCUGUGCCGCA ..........((.....((((((((.((((......))))....(((..(((((((((.(((......((((((.....))))))......)))))))))))).)))))).))))).)). ( -40.30) >consensus AUCUGCAGAAGCCUUUUGCGCACUCAUCACAUUUUCGUGGAAAUCGGAGCGCCAAGGAGCUGUGGAAAGGUGUAUACUCCGCAUCCGUUGCGAAUCCUUGGCGACUCGAACUGUGCCACA ....(((((.....)))))((((...((((......))))...((((..(((((((((((..((((..((.(....).))...))))..))...)))))))))..))))...)))).... (-40.58 = -40.66 + 0.08)

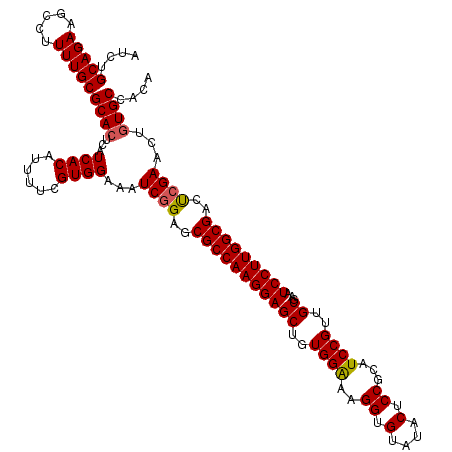
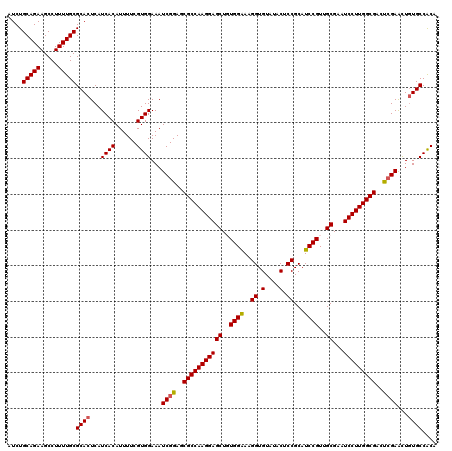
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:26 2006